hmmmmm
1.1 关于 pubmed.mineR
Jyoti Rani, S.Ramachandan and Ab. Rauf Shah (2014). Text mining of PubMed abstracts. R package version 1.0.5
Find it with 'help(package=pubmed.mineR)'
最新版: 1.0.15 , 2019-05-06
从 Title 和 Description 看,“Text Mining of PubMed Abstracts (text and XML) ”,这是一个致力于从 Pubmed Abstarct 文件挖掘文本/数据的包,包里一大堆函数/数据:
ls("package:pubmed.mineR")
# [1] "alias_fn" "altnamesfun"
# [3] "BWI" "cleanabs"
# [5] "cluster_words" "co_occurrence_fn"
# [7] "combineabs" "common_words_new"
# [9] "contextSearch" "cos_sim_calc"
# [11] "cos_sim_calc_boot" "currentabs_fn"
# [13] "Find_conclusion" "find_intro_conc_html"
# [15] "gene_atomization" "genes_BWI"
# [17] "GeneToEntrez" "Genewise"
# [19] "get_gene_sentences" "get_MedlinePlus"
# [21] "get_NMids" "get_original_term"
# [23] "get_original_term2" "get_PMCIDS"
# [25] "get_PMCtable" "get_Sequences"
# [27] "getabs" "getabsT"
# [29] "Give_Sentences" "Give_Sentences_PMC"
# [31] "head_abbrev" "HGNC2UniprotID"
# [33] "HGNCdata" "input_for_find_intro_conc_html"
# [35] "local_uniprotfun" "names_fn"
# [37] "official_fn" "pmids_to_abstracts"
# [39] "previousabs_fn" "prevsymbol_fn"
# [41] "printabs" "pubtator_function"
# [43] "pubtator_result_list_to_table" "readabs"
# [45] "readabsnew" "ready"
# [47] "removeabs" "searchabsL"
# [49] "searchabsT" "sendabs"
# [51] "SentenceToken" "space_quasher"
# [53] "subabs" "subsetabs"
# [55] "tdm_for_lsa" "uniprotfun"
# [57] "whichcluster" "word_associations"
# [59] "word_atomizations" "wordscluster"
# [61] "wordsclusterview" "xmlgene_atomizations"
# [63] "xmlreadabs" "xmlword_atomizations"
# [65] "Yearwise"
2.1 数据准备
在 PubMed 搜索想要的内容,创建 Abstract 文件。
3.1 Text Mining 技能点亮
3.1.1 读取从 PubMed 下载的 Abstract 文件
利用函数 readabs() 读取。
得到的 pubmed_abstracts 是一种 S4 object, Journal , Abstract , PMID 分别是一个 slot.
library(pubmed.mineR)
pubmed_abstracts <- readabs("pubmed_result.txt")
class(pubmed_abstracts)
# [1] "Abstracts"
# attr(,"package")
# [1] "pubmed.mineR"
printabs(pubmed_abstracts) ## 显示开头和结尾部分
3.1.2 从 pubmed_abstracts 提取 PMID
pmid <- pubmed_abstracts@PMID
pmid
# [1] 31158748 31158746 31158660 31157709 31157548 31157505
class(pmid)
# [1] "numeric"
3.1.3 获取注释信息
函数 pubtator_function() 只需输入 PMID(数字或上一步得到的 pmid), 就可以给出相应的信息,如 'Gene’, ’Chemical’, ’Disease’ 等。
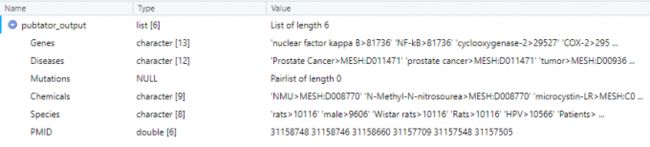
pubtator_output <- pubtator_function(pmid)
也可以直接输入PMID.
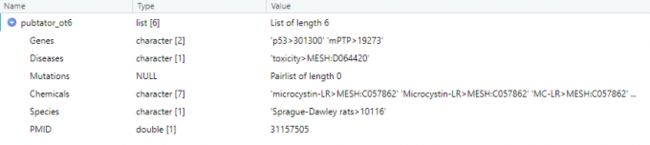
pubtator_ot6 <- pubtator_function(31157505)
然鹅有时……
pubtator_ot1 <- pubtator_function(31158748)
pubtator_ot1
# [1] " No Data "
也有歪果友人发生这样的问题,但直接输入 3.1.2 得到的 pmid 是没问题的ヾ(•ω•`)o
想获取其中一项:
pubtator_output$Genes
# [1] "nuclear factor kappa B>81736" "NF-kB>81736"
# [3] "cyclooxygenase-2>29527" "COX-2>29527"
# [5] "p53>301300" "CTr>116506"
# [7] "NF- kB>81736" "P53>301300"
# [9] "p16>1029" "p53>7157"
# [11] "p53>22060" "p16>13088"
# [13] "mPTP>19273"
pubtator_output$Genes[1]
# [1] "nuclear factor kappa B>81736"
3.1.5 函数SentenceToken() 获取信息
SentenceToken() 可以从 3.1.1 得到的 pubmed_abstracts 中提取语句。
abstractstc <- pubmed_abstracts@Abstract
SentenceToken(abstractstc[1])
# [1] "ahead of print] The synthesis and anticancer activity of 2-styrylquinoline derivatives."
# ……
3.1.6 将获得的信息输出为 .txt 文件
这时要用到这个函数:sendabs()
sendabs(pubmed_abstracts,"mypba.txt")
4.1 一些可视化玩法
4.1.1 统计词频
函数 word_atomizations 可以将整个文本拆分成单词,除去空格、标点符号、常用单词,统计剩下的单词出现的频率。
p53_words <- word_atomizations(pubmed_abstracts)
p53_words[1:10,] ## 只下载了6篇文章的abstract, 样本有点少惹
# words Freq
# 139 cancer 13
# 385 p53 11
# 146 cell 10
# 147 cells 10
# 221 expression 9
# 83 activity 8
# 111 apoptosis 8
# 382 p16 8
# 226 fd 7
# 304 level 7
4.1.2 统计"基因频"
函数 gene_atomization() 基于包内自带的 HGNC 数据库,可以从文本提取 Gene Symbol 和其出现的频率。
p53_gene <- gene_atomization(pubmed_abstracts)
p53_gene
# Gene_symbol Genes Freq
# [1,] "TP53" "tumor protein p53" "2"
# [2,] "ADC" "arginine decarboxylase" "1"
# [3,] "NQO1" "NAD(P)H dehydrogenase, quinone 1" "1"
4.1.3 两个新技能
以 ”关键词“ 和 "年份" 两个参数,得到 PubMed 中相关文章的数量,并可视化。
参考代码来自这里和这里
library(RISmed)
library(dplyr)
library(ggplot2)
library(tidytext)
library(wordcloud)
result <- EUtilsSummary("(p53) AND cancer",
type = "esearch",
db = "pubmed",
datetype = "pdat",
retmax = 10000,
mindate = 1970,
maxdate = 2019)
fetch <- EUtilsGet(result, type = "efetch", db = "pubmed")
abstracts <- data.frame(title = fetch@ArticleTitle,
abstract = fetch@AbstractText,
journal = fetch@Title,
DOI = fetch@PMID,
year = fetch@YearPubmed)
abstracts <- abstracts %>% mutate(abstract = as.character(abstract))
abstracts %>%
group_by(year) %>%
count() %>%
filter(year > 1969) %>%
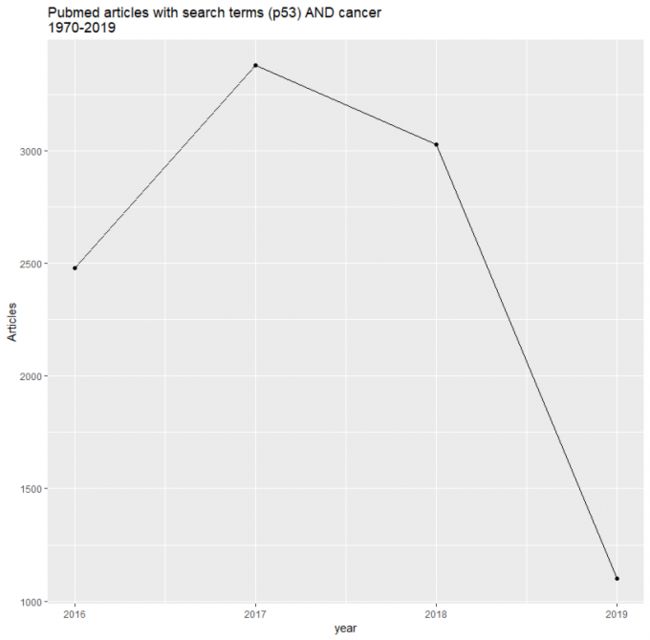
ggplot(aes(year, n)) +
geom_point() +
geom_line() +
labs(title = "Pubmed articles with search terms (p53) AND cancer \n1970-2019", hjust = 0.5,
y = "Articles")
## 然而不知道为什么画出来的图只有2016-2019...(+_+)?
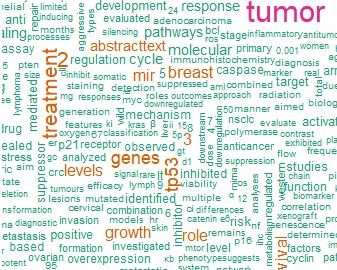
绘制词云图:
cloud <- abstracts %>%
unnest_tokens(word, abstract) %>%
anti_join(stop_words) %>%
count(word, sort = TRUE)
cloud %>%
with(wordcloud(word, n, min.freq = 15, max.words = 500, colors = brewer.pal(8, "Dark2")), scale = c(8,.3), per.rot = 0.4)
得到了一张又方又丑的图。(:з)∠)_
References
Zipf's Law: Pubmed.mineR: text mining from the biomedical literature with the R programming language https://zipfslaw.org/2015/10/19/pubmed-miner/
pubmed.mineR: Breaking the ice - Beginners tutorial http://ramuigib.blogspot.com/2018/07/breaking-ice-beginners-tutorial.html
PHARMACOVIGILANCE ANALYTICS: Mining PubMed for Drug Induced Acute Kidney Injury http://www.pharmacovigilanceanalytics.com/methods/text-mining/mining-pubmed-for-drug-induced-acute-kidney-injury/
rstudio-pubs-static: Pubmed2: searching Pubmed for articles on public health and data science https://rstudio-pubs-static.s3.amazonaws.com/235239_f75d5005bc0f4015bd97fb4be182144a.html#
最后,向大家隆重推荐生信技能树的一系列干货!
- 生信技能树全球公益巡讲:https://mp.weixin.qq.com/s/E9ykuIbc-2Ja9HOY0bn_6g
- B站公益74小时生信工程师教学视频合辑:https://mp.weixin.qq.com/s/IyFK7l_WBAiUgqQi8O7Hxw
- 招学徒:https://mp.weixin.qq.com/s/KgbilzXnFjbKKunuw7NVfw