想让这张图当封面
1.1 关于 easyPubMed
作者是 Damiano Fantini ,2019-03-29 发表,甩链接。
看它的 Title: 搜索和读取 PubMed 上的文章发表信息。
看它的自我介绍: easyPubMed 可以查询 NCBI Entrez,以 XML 或 文本 格式获得 PubMed 信息,可以提取、整合数据,可以 轻 而 易 举 地下载一大堆记录信息,比如单独得到 作者、单位、题目、关键词、摘要、发表时间……
2.1 包里的函数
一共12个:
articles_to_list , article_to_df , batch_pubmed_download , custom_grep , EPMsamples, fetch_all_pubmed_ids , fetch_pubmed_data . get_pubmed_ids ,get_pubmed_ids_by_fulltitle , PubMed_stopwords , table_articles_byAuth ,trim_address
3.1 小白使用教程
第一步当然是:
library(easyPubMed)
再加载一些要用到的包:
library(XML)
library(dplyr)
library(kableExtra)
library(parallel)
library(foreach)
library(doParallel)
3.1.1 简单的数据获取
以这个包的作者为例,获取TA的所有文章,[AU]-author.
query <- "Damiano Fantini[AU]"
用 get_pubmed_ids() 得到一个 list 文件,包含后续操作需要的信息。
A PubMed query by the
get_pubmed_ids()function results in:
- the query results are posted on the Entrez History Server ready for retrieval
- the function returns a list containing all information to access and download resuts from the server
entrez_id <- get_pubmed_ids(query)
用 fetch_pubmed_data() 得到 PubMed 数据。
abstracts_txt <- fetch_pubmed_data(entrez_id, format = "abstract")
print(abstracts_txt[1:16])
这里的 "format" 允许这些选项:"asn.1", "xml", "medline", "uilist", "abstract"
还可以用 fetch_pubmed_data() 结合 XML包,得到所有文章的标题。
abstracts_xml <- fetch_pubmed_data(entrez_id,format = "xml")
class(abstracts_xml)
示例得到的结果是:
## [1] "XMLInternalDocument" "XMLAbstractDocument"然后就能愉快地继续下面的操作:
my_titles <- unlist(xpathApply(my_abstracts_xml, "//ArticleTitle", saveXML)) my_titles <- gsub("(^.{5,10}Title>)|(<\\/.*$)", "", my_titles) my_titles[nchar(my_titles)>75] <- paste(substr(my_titles[nchar(my_titles)>75], 1, 70), "...", sep = "") print(my_titles)最后愉快地得到所有文章的 title.
然鹅……我得到的结果是:
abstracts_xml <- fetch_pubmed_data(entrez_id,format = "xml")
## 默认的format就是"xml",这个可以不输
class(abstracts_xml)
## [1] "character"
titles <- unlist(xpathApply(abstracts_xml, "//ArticleTitle", saveXML))
## Error in UseMethod("xpathApply") :
no applicable method for 'xpathApply' applied to an object of class "character"
绕路用另一种方法,函数 custom_grep 也可以获取所有文章的 title.
titles <- custom_grep(abstracts_xml, "ArticleTitle", "char")
## format,c("list", "char"):
print(titles)
成功!
3.1.2 以 TXT 或 XML 格式下载并保存信息
通过 batch_pubmed_download() 将数据保存为 txt 或 xml 文件。
## 搜索标题里有 APE1 或 OGG1 这两个基因——在2012-2016年间发表的文章
new_query <- "(APE1[TI] OR OGG1[TI]) AND (2012[PDAT]:2016[PDAT])"
## 设置输出文件格式、文件名前缀
outfile <- batch_pubmed_download(pubmed_query_string = new_query,
format = "xml",
batch_size = 150,
dest_file_prefix = "easyPM_example")
outfile
## [1] "easyPM_example01.txt" "easyPM_example02.txt"
不造为什么得到的永远是txt……作者的示例里输出的是:
## [1] "easyPM_example01.xml" "easyPM_example02.xml"
3.1.3 从单独的 PubMed 记录里提取信息
custom_grep 函数可以将 XML 转换为字符串,从特定的 PubMed 记录中提取相关信息,返回 list 或 character.
PM_list <- articles_to_list(abstracts_xml)
## 任意选其中的一条
custom_grep(PM_list[13], tag = "DateCompleted")
## [[1]]
## [1] " 2011 01 31 "
但要注意:被选择的那条 record 是否含有要提取的 tag.
custom_grep(PM_list[8], tag = "DateCompleted")
## list()
另一个 tag 的例子:
custom_grep(PM_list[13], tag = "LastName", format = "char")
## [1] "Fantini" "Vascotto" "Marasco" "D'Ambrosio" "Romanello" "Vitagliano"
## [7] "Pedone" "Poletto" "Cesaratto" "Quadrifoglio" "Scaloni" "Radicella"
## [13] "Tell"
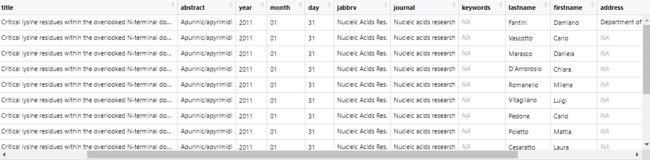
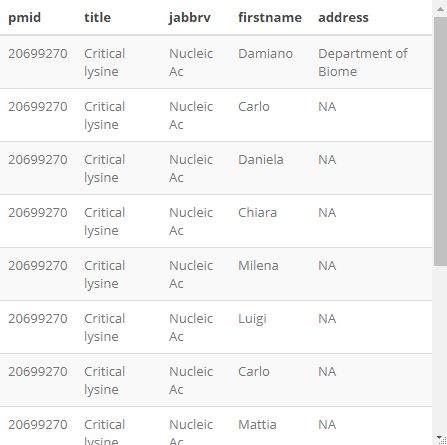
函数 article_to_df() 可以将字符串转换为 data.frame, 尤其是由 articles_to_list 得到的元素,得到的 data.frame 的列名为 "pmid", "doi", "title", "abstract", "year", "month", "day", "jabbrv", "journal", "lastname", "firstname", "address", "email".
df <- article_to_df(PM_list[13], max_chars = 18)
colnames(df)
## [1] "pmid" "doi" "title" "abstract" "year" "month" "day"
## [8] "jabbrv" "journal" "keywords" "lastname" "firstname" "address" "email"
调整表格:
df$title <- substr(df$title, 1, 15)
df$address <- substr(df$address, 1, 19)
df$jabbrv <- substr(df$jabbrv, 1, 10)
df[,c("pmid", "title", "jabbrv", "firstname", "address")] %>% kable() %>%
kable_styling(bootstrap_options = 'striped')
如果多个作者有相同的单位信息,设置参数 autofill 为 “TRUE”.
df2 <- article_to_df(PM_list[13], autofill = TRUE)
df2$title <- substr(df2$title, 1, 15)
df2$address <- substr(df2$address, 1, 19)
df2$jabbrv <- substr(df2$jabbrv, 1, 10)
df2[,c("pmid", "title", "jabbrv", "firstname", "address")] %>% kable() %>%
kable_styling(bootstrap_options = 'striped')
3.1.4 从 XML PubMed 记录中自动提取数据
函数 table_articles_byAuth() 可以迅速从多个 XML 记录获得作者信心和文章发表数据,该函数包含5个参数:
- pubmed_data: an XML file or an XML object with PubMed records
- max_chars and autofill: same as discussed in the previous example
- included_authors: one of the following options c(“first”, “last”, “all”). The function can return data corresponding to the first, the last or all the authors for each PubMed record.
- dest_file: if not NULL, the function attempts writing its output to the selected file. Existing files will be overwritten.
new_PM_query <- "(APEX1[TI] OR OGG1[TI]) AND (2010[PDAT]:2013[PDAT])"
outfile2 <- batch_pubmed_download(pubmed_query_string = new_PM_query, dest_file_prefix = "apex1_sample")
## [1] "PubMed data batch 1 / 1 downloaded..."
new_PM_file <- outfile2[1]
new_PM_df <- table_articles_byAuth(pubmed_data = new_PM_file, included_authors = "first", max_chars = 0)
## Processing PubMed data .................................................. done!
转换为 data.frame , 调整表格。
new_PM_df$address <- substr(new_PM_df$address, 1, 28)
new_PM_df$jabbrv <- substr(new_PM_df$jabbrv, 1, 9)
new_PM_df[1:10, c("pmid", "year", "jabbrv", "lastname", "address")] %>%
kable() %>% kable_styling(bootstrap_options = 'striped')
4.1 老司机进阶
4.1.1 准备数据
my_query <- 'Damiano Fantini[AU] AND '
my_query <- get_pubmed_ids(my_query)
my_batches <- seq(from = 1, to = my_query$Count, by = 10)
my_abstracts_xml <- lapply(my_batches, function(i) {
fetch_pubmed_data(my_query, retmax = 1000, retstart = i)
})
## 储存为 list
all_xml <- list()
for(x in my_abstracts_xml) {
xx <- articles_to_list(x)
for(y in xx) {
all_xml[[(1 + length(all_xml))]] <- y
}
}
4.1.2 快速提取 PMID, 标题,摘要
利用参数 article_to_df(, getAuthors = FALSE) ,省去作者信息,可以快速得到其他全部数据。
## 对整个过程计时
t.start <- Sys.time()
## max_chars = -1 即提取全部摘要
final_df <- do.call(rbind, lapply(all_xml, article_to_df,
max_chars = -1, getAuthors = FALSE))
t.stop <- Sys.time()
print(t.stop - t.start)
## Time difference of 0.4308472 secs
选择感兴趣的项目并呈现出表格:
interested_df <- final_df[,c("pmid","title","year","jabbrv")]
interested_df %>%
head(n=4) %>% kable() %>% kable_styling(bootstrap_options = 'striped')
4.1.3 提取全部信息,包括关键词
利用参数 article_to_df(, getKeywords = TRUE) 得到文章关键词。
t.start <- Sys.time()
keyword_df <- do.call(rbind, lapply(all_xml,
article_to_df, autofill = T,
max_chars = 100, getKeywords = T))
t.stop <- Sys.time()
print(t.stop - t.start)
## Time difference of 4.216758 secs
print(keyword_df$keywords[seq(1, 150, by = 15)])
4.1.4 将任务拆分,多任务平行提取数据
keyword_df[seq(1, 100, by = 10), c("lastname", "keywords", "abstract")] %>%
kable() %>% kable_styling(bootstrap_options = 'striped')
作者在这里注释:
Load required packages (available from CRAN).
This will work on UNIX/LINUX systems.
Windows systems may not support the following code.
然鹅用win的我并没有发现有什么异常、以及 和前面的 区 别(⊙ˍ⊙)
4.1.5 利用 NCBI/Entrez API key 实现更快的信息获取
没有 API Key, 所以以下均为 复制:
# define a PubMed Query: this should return 40 results
my_query <- '"immune checkpoint" AND 2010[DP]:2012[DP]'
# Monitor time, and proceed with record download -- USING API_KEY!
t_key1 <- Sys.time()
set_01 <- batch_pubmed_download(my_query,
api_key = "NNNNNNNNNNe9108aee96ace507af23a4eb09",
batch_size = 2, dest_file_prefix = "TMP_api_")
t_key2 <- Sys.time()
# Monitor time, and proceed with record download -- DO NOT USE API_KEY!
t_nok1 <- Sys.time()
set_02 <- batch_pubmed_download(my_query,
batch_size = 2, dest_file_prefix = "TMP_no_")
t_nok2 <- Sys.time()
# Compute time differences
# The use of a key makes the process faster
print(paste("With key:", t_key2 - t_key1))
## [1] "With key: 20.7291417121887"
print(paste("W/o key:", t_nok2 - t_nok1))
## [1] "W/o key: 24.6694004535675"
总之就是用这个方法超快
4.1.6 文章全长标题的精准匹配
2.11 版的新函数 get_pubmed_ids_by_fulltitle(),可以直接输入文章标题,再敲几行代码得到想要的信息。
试一下 Boss Jimmy 布置的作业:
ftitle_query <- "Identification of hub genes and outcome in colon cancer based on bioinformatics analysis"
my_field <- "[Title]"
fullti <- get_pubmed_ids_by_fulltitle(ftitle_query, field = my_field)
print(as.numeric(fullti$IdList$Id[1]))
## [1] 30643458
就这样,得到了这篇文章的 PMID.
References
- easyPubMed official website including news, vignettes, and further information https://www.data-pulse.com/dev_site/easypubmed/
最后,向大家隆重推荐生信技能树的一系列干货!
- 生信技能树全球公益巡讲:https://mp.weixin.qq.com/s/E9ykuIbc-2Ja9HOY0bn_6g
- B站公益74小时生信工程师教学视频合辑:https://mp.weixin.qq.com/s/IyFK7l_WBAiUgqQi8O7Hxw
- 招学徒:https://mp.weixin.qq.com/s/KgbilzXnFjbKKunuw7NVfw