原文地址:https://mp.weixin.qq.com/s/AK3UICxBYTnG98jy_6q4gA
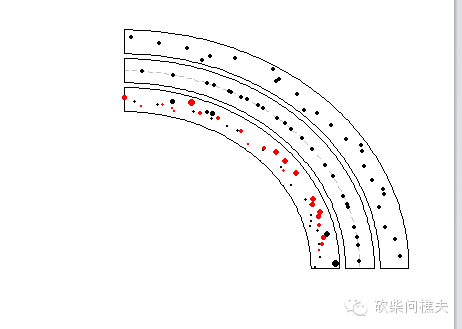
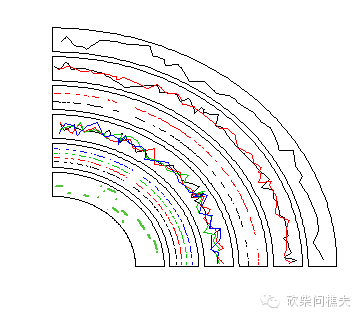
我们首先看一下绘制点:
par(mar = c(1, 1, 1, 1))
set.seed(1)
#起始角度90
circos.par("track.height" = 0.1, start.degree = 90,gap.degree=270)
#初始化(绘制第一染色体,表示仅有一个扇形)
circos.initializeWithIdeogram(chromosome.index = "chr1", plotType = NULL)
#构造数据框
bed = generateRandomBed(nr = 300)
#创建轨道,并在上面添加点
circos.genomicTrackPlotRegion(bed, panel.fun = function(region, value, ...) {
circos.genomicPoints(region, value, pch = 16, cex = 0.5, ...)
})
#继续构造数据集
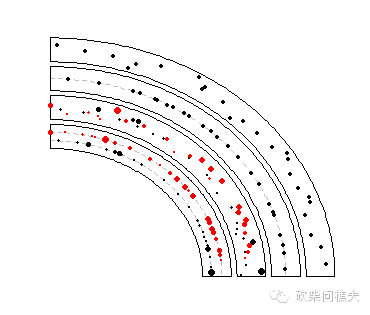
bed = generateRandomBed(nr = 300)
#堆叠(类似与ggplot中的位置参数stack),点上加线
circos.genomicTrackPlotRegion(bed, stack = TRUE,
panel.fun = function(region, value, ...) {
circos.genomicPoints(region, value, pch = 16, cex = 0.5, ...)
#在当前数据框
i = getI(...)
cell.xlim = get.cell.meta.data("cell.xlim")
circos.lines(cell.xlim, c(i, i), lty = 2, col = "#00000040")
})
#产生两个数据集
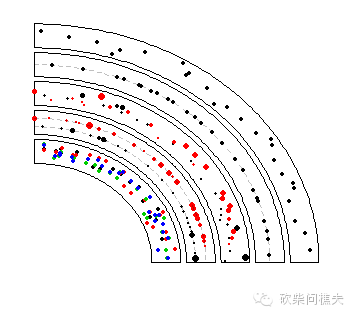
bed1 = generateRandomBed(nr = 300)
bed2 = generateRandomBed(nr = 300)
#组合在一起
bed_list = list(bed1, bed2)
#两个数据集在同一个轨道上添加点(i=get(...)必须有)
circos.genomicTrackPlotRegion(bed_list,
panel.fun = function(region, value, ...) {
cex = (value[[1]] - min(value[[1]]))/(max(value[[1]]) - min(value[[1]]))
i = getI(...)
circos.genomicPoints(region, value, cex = cex, pch = 16, col = i, ...)
})
#堆叠circos.genomicTrackPlotRegion(bed_list, stack = TRUE,
panel.fun = function(region, value, ...) {
cex = (value[[1]] - min(value[[1]]))/(max(value[[1]]) - min(value[[1]]))
i = getI(...)
circos.genomicPoints(region, value, cex = cex, pch = 16, col = i, ...)
cell.xlim = get.cell.meta.data("cell.xlim")
circos.lines(cell.xlim, c(i, i), lty = 2, col = "#00000040")
})
#构造数据集(多个变量)
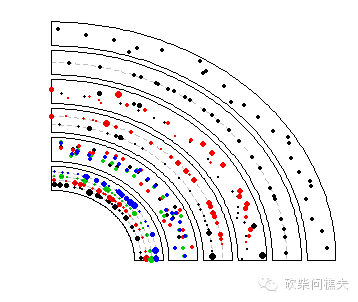
bed = generateRandomBed(nr = 300, nc = 4)
circos.genomicTrackPlotRegion(bed,
panel.fun = function(region, value, ...) {
circos.genomicPoints(region, value, cex = 0.5, pch = 16, col = 1:4, ...)
})
bed = generateRandomBed(nr = 300, nc = 4)
circos.genomicTrackPlotRegion(bed, stack = TRUE,
panel.fun = function(region, value, ...) {
cex = (value[[1]] - min(value[[1]]))/(max(value[[1]]) - min(value[[1]]))
i = getI(...)
circos.genomicPoints(region, value, cex = cex, pch = 16, col = i, ...)
cell.xlim = get.cell.meta.data("cell.xlim")
circos.lines(cell.xlim, c(i, i), lty = 2, col = "#00000040")
})
circos.clear()
下面绘制线:
par(mar = c(1, 1, 1, 1))
circos.par("track.height" = 0.1, start.degree = 90, gap.degree = 270)
circos.initializeWithIdeogram(chromosome.index = "chr1", plotType = NULL)
bed = generateRandomBed(nr = 500)
circos.genomicTrackPlotRegion(bed,
panel.fun = function(region, value, ...) {
circos.genomicLines(region, value, type = "l", ...)
})
bed1 = generateRandomBed(nr = 500)
bed2 = generateRandomBed(nr = 500)
bed_list = list(bed1, bed2)
circos.genomicTrackPlotRegion(bed_list,
panel.fun = function(region, value, ...) {
i = getI(...)
circos.genomicLines(region, value, col = i, ...)
})
circos.genomicTrackPlotRegion(bed_list, stack = TRUE,
panel.fun = function(region, value, ...) {
i = getI(...)
circos.genomicLines(region, value, col = i, ...)
})
bed = generateRandomBed(nr = 500, nc = 4)
circos.genomicTrackPlotRegion(bed,
panel.fun = function(region, value, ...) {
circos.genomicLines(region, value, col = 1:4, ...)
})
bed = generateRandomBed(nr = 500, nc = 4)
circos.genomicTrackPlotRegion(bed, stack = TRUE,
panel.fun = function(region, value, ...) {
i = getI(...)
circos.genomicLines(region, value, col = i, ...)
})
bed = generateRandomBed(nr = 200)
circos.genomicTrackPlotRegion(bed,
panel.fun = function(region, value, ...) {
circos.genomicLines(region, value, type = "segment", lwd = 2,
col = rand_color(nrow(region)), ...)
})
circos.clear()
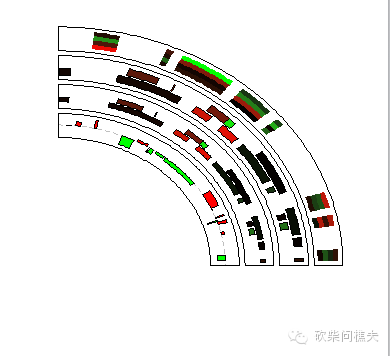
绘制矩形:
par(mar = c(1, 1, 1, 1))
circos.par("track.height" = 0.1, start.degree = 90,gap.degree = 270)
circos.initializeWithIdeogram(chromosome.index = "chr1", plotType = NULL)
f = colorRamp2(breaks = c(-1, 0, 1), colors = c("green", "black", "red"))
bed = generateRandomBed(nr = 100, nc = 4)
circos.genomicTrackPlotRegion(bed, stack = TRUE,
panel.fun = function(region, value, ...) {
circos.genomicRect(region, value, col = f(value[[1]]), border = NA, ...)
})
bed1 = generateRandomBed(nr = 100)
bed2 = generateRandomBed(nr = 100)
bed_list = list(bed1, bed2)
circos.genomicTrackPlotRegion(bed_list, stack = TRUE,
panel.fun = function(region, value, ...) {
i = getI(...)
circos.genomicRect(region, value, ytop = i + 0.4, ybottom = i - 0.4,
col = f(value[[1]]), ...)
})
circos.genomicTrackPlotRegion(bed_list, ylim = c(0, 3),
panel.fun = function(region, value, ...) {
i = getI(...)
circos.genomicRect(region, value, ytop = i + 0.4, ybottom = i - 0.4,
col = f(value[[1]]), ...)
})
bed = generateRandomBed(nr = 200)
circos.genomicTrackPlotRegion(bed,
panel.fun = function(region, value, ...) {
circos.genomicRect(region, value, ytop.column = 1, ybottom = 0,
col = ifelse(value[[1]] > 0, "red", "green"), ...)
cell.xlim = get.cell.meta.data("cell.xlim")
circos.lines(cell.xlim, c(0, 0), lty = 2, col = "#00000040")
})
circos.clear()
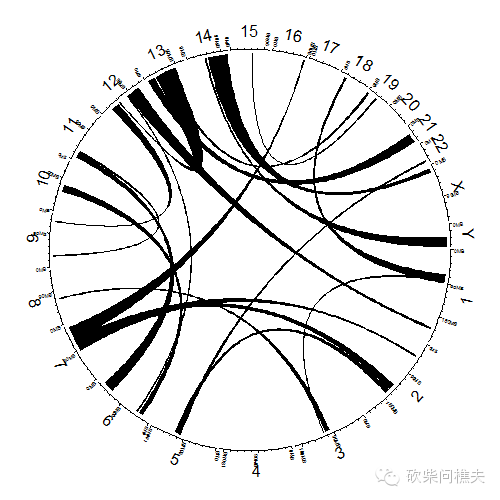
点到点,点到区间,区间到区间的连线:
#构造数据
bed1 = generateRandomBed(nr = 100)
#抽提部分数据
bed1 = bed1[sample(nrow(bed1), 20), ]
bed2 = generateRandomBed(nr = 100)
bed2 = bed2[sample(nrow(bed2), 20), ]
#绘制轨道
circos.initializeWithIdeogram(plotType = c("axis", "labels"))
#区间到区间的连线
circos.genomicLink(bed1, bed2)
circos.clear()
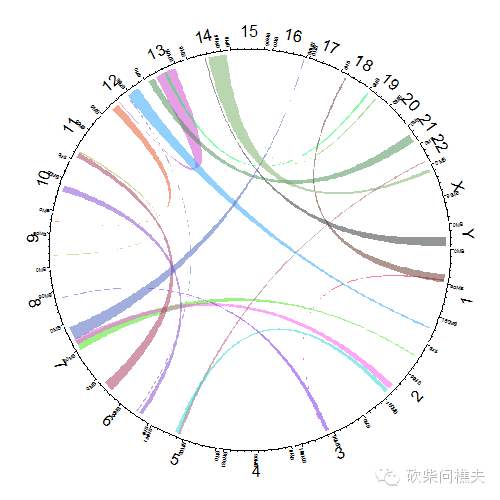
#我们也是可以着色的
circos.initializeWithIdeogram(plotType = c("axis", "labels"))
circos.genomicLink(bed1, bed2, col = rand_color(nrow(bed1), transparency = 0.5), border = NA)
我们在基因组上也是可以高亮染色体信息的:
circos.par("track.height" = 0.1)
circos.initializeWithIdeogram(plotType = c("axis", "labels"))
#绘制5条轨道
for(i in 1:5) {
bed = generateRandomBed(nr = 100)
circos.genomicTrackPlotRegion(bed, panel.fun = function(region, value, ...) {
circos.genomicPoints(region, value, pch = 16, cex = 0.5, ...)
})
}
#高亮染色体
highlight.chromosome(c("chrX", "chrY", "chr1"))
highlight.chromosome("chr3", col = "#00FF0040", padding = c(0.05, 0.05, 0.15, 0.05))
highlight.chromosome("chr5", col = NA, border = "red", lwd = 2, padding = c(0.05, 0.05, 0.15, 0.05))
highlight.chromosome("chr7", col = "#0000FF40", track.index = c(2, 4, 5))
highlight.chromosome(c("chr9", "chr10", "chr11"), col = NA, border = "green",lwd = 2, track.index = c(2, 4, 5))
highlight.chromosome(paste0("chr", c(1:22, "X", "Y")), col = "#FFFF0040", track.index = 6)
circos.clear()
更多circlize信息,请看帮助文档……