序列比对和序列特征分析总目录
网址:https://blast.ncbi.nlm.nih.gov/Blast.cgi
运行方式:本地或web
基本的BLAST工具包括:
blastn:核酸搜核酸数据库
blastp:蛋白质搜蛋白质数据库
blastx:DNA用所有可能的阅读框翻译成翻译成蛋白后搜蛋白数据库
tblastn:查询的蛋白序列搜索核酸数据库中,DNA序列翻译后的蛋白序列
tblastx:核酸序列翻译成蛋白质后搜索核酸数据库中的核酸序列翻译后的蛋白质序列。也就是查询的蛋白和数据库中的DNA都翻译成蛋白进行比对。
一: web blast
举一个例子说明
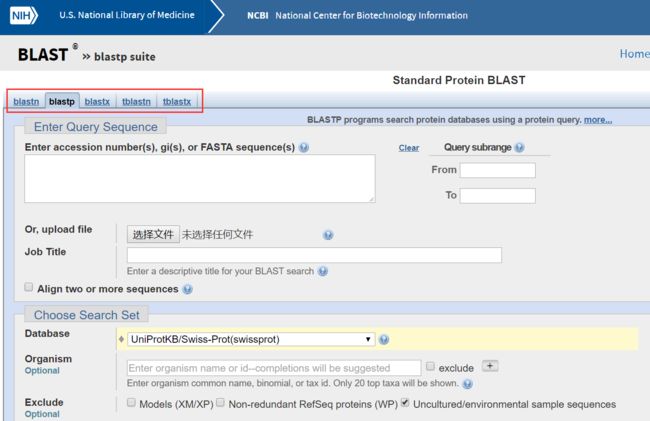
图1可以看到,输入框可以输入accesion number,gi,或FASTA序列,也可以上传文件。
job title给查询的任务取个名字。
参数设置
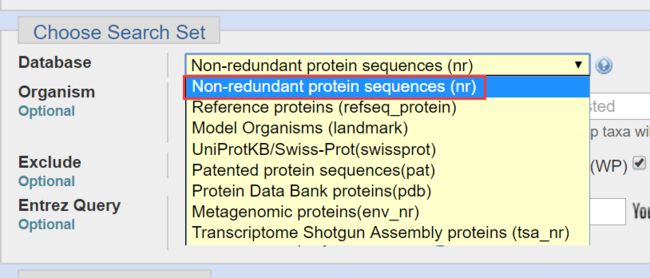
- Database:图2,一般选择nr,即非冗余蛋白序列数据库,该库包括GenBank CDS tranlations,RefSSeq Proteins,PDB,Swiss-Prot,PIR和PRF全体数据库的非冗余数据
- 算法参数设置
首先每个参数后面都有说明,可以详细查看该选择哪个 - Organism可以限制物种
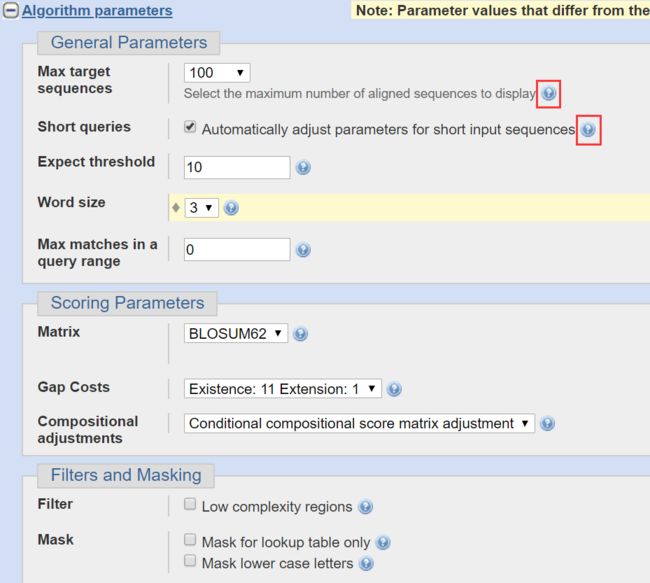
- Expect threshold期望阈值,默认10
- word size字长,默认3,还可以设置为10或2,数值小搜索的结果会增加,速度会变慢
- matrix序列比对的打分矩阵,默认LOSUM62
-
Gap costs:BLAST采取线性空间罚分方式,为开放罚分和延伸罚分,默认是开放罚分值11,延伸罚分值1
结果解读
- 搜索详细情况描述。图4,查询的分子类型,比对的数据库,都有描述。
-
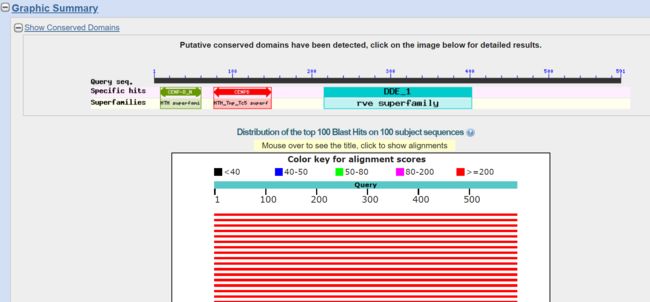
图形结果。查询序列含有的保守结构域,以及数据库中与查询序列匹配项的图形。不同彩色条带颜色代表得分的高低。
-
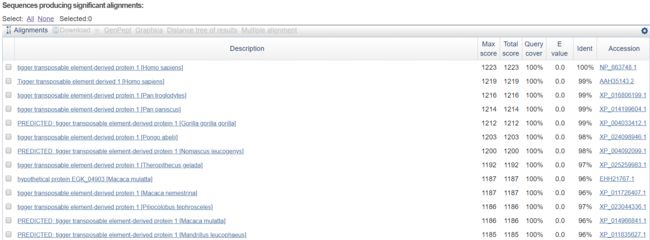
详细列表信息.与查询的序列匹配的数据库中的序列列表,每一个序列包括score,evalue,identity,accesion等。
-
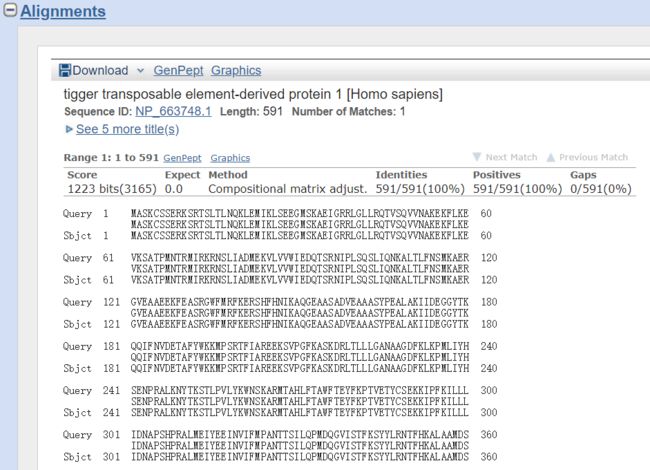
查询序列与数据库中的匹配序列之间的双序列比对情况。包括score,expect,identity同一性得分,positive相似性分值,gaps空位。
总结:
web版的blast方便,快捷,容易操作,数据库更新快。确定是不利于操作大力量数据,也不能自定义搜索的数据库,只能对NCBI提供的数据库进行序列相似性分析。所以
NCBI提供了本地化安装的blast软件包,这样就可以构建自己的数据库,提高同源性分析的准确性和一致性。
二: LINUX下BLAST的安装与运行
优点:速度快,灵活性大,可自己配置库
缺点:序列数据库下载量大,并且更新麻烦,需要重新下载
1 安装配置BLAST
1.1 利用conda安装,关于conda请看之前的简文
#启动环境
$ source ~/miniconda3/bin/activate
$ conda install blast
比较简单
1.2 直接下载安装
首先在ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/下载最新版本的BLAST程序。
wget ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/LATEST/ncbi-blast-2.8.1+-x64-linux.tar.gz
Connecting to ftp.ncbi.nlm.nih.gov (ftp.ncbi.nlm.nih.gov)|130.14.250.12|:21... connected.
Logging in as anonymous ... Logged in!
==> SYST ... done. ==> PWD ... done.
==> TYPE I ... done. ==> CWD (1) /blast/executables/LATEST ... done.
==> SIZE ncbi-blast-2.8.1+-x64-linux.tar.gz ... 241992963
==> PASV ... done. ==> REST 173905320 ... done.
==> RETR ncbi-blast-2.8.1+-x64-linux.tar.gz ... done.
Length: 241992963 (231M), 68087643 (65M) remaining (unauthoritative)
ncbi-blast-2.8.1+-x64-linux.tar. 100%[++++++++++++++++++++++++++++++++++++++++================>] 230.78M 2.92MB/s in 33s
2019-01-23 13:30:52 (1.98 MB/s) - ‘ncbi-blast-2.8.1+-x64-linux.tar.gz’ saved [241992963]
接下来解压缩
$ tar -xzvf ncbi-blast-2.8.1+-x64-linux.tar.gz
$ rm ncbi-blast-2.8.1+-x64-linux.tar.gz
$ mv ncbi-blast-2.8.1+/ blast
$ cd blast
$ cd bin
$ ls
可执行文件显示如下
blastdb_aliastool blastn deltablast makeblastdb rpsblast tblastx
blastdbcheck blastp dustmasker makembindex rpstblastn update_blastdb.pl
blastdbcmd blastx get_species_taxids.sh makeprofiledb segmasker windowmasker
blast_formatter convert2blastmask legacy_blast.pl psiblast tblastn
2 运行
要进行序列比对,得有以下几个条件
第一,有查询序列,并有特定格式
第二,有目标序列库,蛋白库还是DNA库
第三,确定查询工具,blastn,blastp,blastx,tblastx,tblastn
第四,设定合适参数开始运行
具体用法BLAST手册《BLAST Command Line Applications User Manual》
2.1本地建库
第1️⃣:NCBI下载nt和nr库文件到本地
BLAST database
获取blast database的最好方法是NCBI下载。
通过运行$ update_blastdb.pl --decompress nr [*]程序,可以下载预先格式化的NCBI BLAST database。
#先创建blast_db目录
~$ mkdir blast_db
$ cd blast_db
# 耗时很长,放入后台
$ nohup time update_blastdb.pl nt nr > log &
$ nohup time tar -zxvf *.tar.gz > log2 &
说明:nt为核酸,nr为蛋白质
监控库文件是否下载完成,如何判断? 1. 查看log文件是否有提示;2. 查看update_blastdb.pl是否还在运行:执行ps -aef | grep update_blastdb.pl | grep -v update_blastdb.pl 命令,如过没有结果,则说明没有运行了。
这部分来自hoptop也可以是从ncbi上直接下载一系列nt/nr.xx.tar.gz文件,然后解压缩即可,后续可以用update_blastdb.pl进行数据更新。
报错: 使用update_blastdb.pl更新和下载数据库时候出现模块未安装的问题。解决方法,首先用conda安装对应的模块,然后修改update_blastdb.pl的第一行,即shebang部分,以conda的perl替换,或者按照如下方法执行。
perlwhich update_blastdb.pl
下载过程中请确保网络状态良好,否则会出现Downloading nt.00.tar.gz...Unable to close datastream报错。
第2️⃣:用自己的序列建库makeblastdb
BLAST需要进行序列数据的索引格式化,然后才可以进行序列的比对搜索,所以先用makeblastdb进行格式化
makeblastdb -in mydb.fasta -dbtype nucl -parse_seqids -out dbname
简单解释如下:
-in :表示输入的数据文件
-dbtype:序列数据类型,核酸nucl,蛋白质prot
详细用法:
$ makeblastdb -help
USAGE
makeblastdb [-h] [-help] [-in input_file] [-input_type type]
-dbtype molecule_type [-title database_title] [-parse_seqids]
[-hash_index] [-mask_data mask_data_files] [-mask_id mask_algo_ids]
[-mask_desc mask_algo_descriptions] [-gi_mask]
[-gi_mask_name gi_based_mask_names] [-out database_name]
[-max_file_sz number_of_bytes] [-logfile File_Name] [-taxid TaxID]
[-taxid_map TaxIDMapFile] [-version]
DESCRIPTION
Application to create BLAST databases, version 2.7.1+
REQUIRED ARGUMENTS
-dbtype
Molecule type of target db
OPTIONAL ARGUMENTS
-h
Print USAGE and DESCRIPTION; ignore all other parameters
-help
Print USAGE, DESCRIPTION and ARGUMENTS; ignore all other parameters
-version
Print version number; ignore other arguments
*** Input options
-in
Input file/database name
Default = `-'
-input_type
Type of the data specified in input_file
Default = `fasta'
*** Configuration options
-title
Title for BLAST database
Default = input file name provided to -in argument
所需的基因组文件可以下载,也可以是自己测序的。现在我用我自己的文件mybp.fa,看如何建库
$ makeblastdb -in mybp.fa -dbtype nucl -out mybp -parse_seqids
显示如下,建库完成
Building a new DB, current time: 01/24/2019 07:55:54
New DB name: /mnt/d/yinlibioinfor/mybp
New DB title: mybp.fa
Sequence type: Nucleotide
Keep MBits: T
Maximum file size: 1000000000B
Adding sequences from FASTA; added 2 sequences in 0.0724082 seconds.
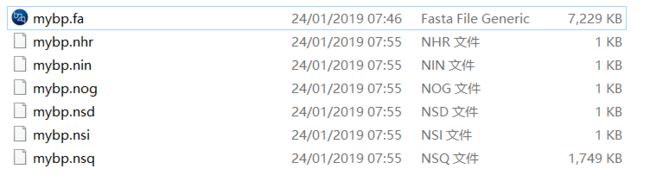
本地目标文件夹会产生以下几个数据库文件
2.2:运行
最基本的BLAST命令包含query和db参数
blastn -db mydb.fa -query xx.fa -out results.out
- query: 检索文件
- remote:上面已经把nt nr下载到本地,如果不下载可以加 -remote,速度会慢
-evalue,默认10,设置输出结果的e-value。如果E小于10-5,说明两条序列有较高同源性,统计学意义显著。若小于10-6则表示同源性非常高。几乎可以百分百确定。
A:用我自己的一段序列查询
$ blastn -db mybp -query query.fa
结果如下:
Database: mybp.fa
2 sequences; 7,163,595 total letters
Query= query
Length=366
Score E
Sequences producing significant alignments: (Bits) Value
Chromosome2 circular 676 0.0
>Chromosome2 circular
Length=3363129
Score = 676 bits (366), Expect = 0.0
Identities = 366/366 (100%), Gaps = 0/366 (0%)
Strand=Plus/Plus
Query 1 ATGATCAAGGACGTTCTACGACTTAAATTCGACGGCAGCCTTTCGCACGATCGGATCGCC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 1167368 ATGATCAAGGACGTTCTACGACTTAAATTCGACGGCAGCCTTTCGCACGATCGGATCGCC 1167427
Query 61 ACGTCGCTGGGCATTTCCAAAAGCGTGGTCACGAAGCACGTCGGACCGGCGGGCGCCGCC 120
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 1167428 ACGTCGCTGGGCATTTCCAAAAGCGTGGTCACGAAGCACGTCGGACCGGCGGGCGCCGCC 1167487
Query 121 GGGCTGGACCGGGCAAGCACCTGCGAGATGGACGAGGGCGAGCGCAAGCGGCGGCTACTC 180
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 1167488 GGGCTGGACCGGGCAAGCACCTGCGAGATGGACGAGGGCGAGCGCAAGCGGCGGCTACTC 1167547
Query 181 GGCAAGCCCATGAGACCAGCGACCTACGTCCAGCCCGATTACGGGCGCATCCATCAGGAA 240
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 1167548 GGCAAGCCCATGAGACCAGCGACCTACGTCCAGCCCGATTACGGGCGCATCCATCAGGAA 1167607
Query 241 CTGCGCCGCAAAGGCGTGACGTTGACGCTGCTGTGGGAGGAGTACCAAGTCGAGTTCGCC 300
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 1167608 CTGCGCCGCAAAGGCGTGACGTTGACGCTGCTGTGGGAGGAGTACCAAGTCGAGTTCGCC 1167667
Query 301 GGCCGGCAAACCTACCGCTCTACGCGCAATTCTGCGAGCACTACAAGGCGTTCACAAAGC 360
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 1167668 GGCCGGCAAACCTACCGCTCTACGCGCAATTCTGCGAGCACTACAAGGCGTTCACAAAGC 1167727
Query 361 GTCTGA 366
||||||
Sbjct 1167728 GTCTGA 1167733
query序列本身就是来自上述基因组,所以完全匹配。
B:使用远程服务器在线比对,但是速度真的是很慢
$ blastn -db nr -remote -query query.fa
小部分结果如下
Database: Nucleotide collection (nt)
49,985,097 sequences; 191,944,857,236 total letters
Query= query
Length=366
RID: 4J1C2RW4015
Score E
Sequences producing significant alignments: (Bits) Value
CP033704.1 Burkholderia pseudomallei strain FDAARGOS_593 chromos... 676 0.0
CP033701.1 Burkholderia pseudomallei strain FDAARGOS_594 chromos... 676 0.0
CP025307.1 Burkholderia pseudomallei strain D286 chromosome 2, c... 676 0.0
CP025305.1 Burkholderia pseudomallei strain R15 chromosome R15.2... 676 0.0
CP025303.1 Burkholderia pseudomallei strain PMC2000 chromosome P... 676 0.0
CP025301.1 Burkholderia pseudomallei strain H10 chromosome H10.2 676 0.0
CP019043.1 Burkholderia pseudomallei strain 14M0960418 chromosom... 676 0.0
CP012577.1 Burkholderia pseudomallei strain 982 chromosome 2, co... 676 0.0
CP012093.1 Burkholderia pseudomallei strain 350105 chromosome 2 ... 676 0.0
CP009297.1 Burkholderia pseudomallei 406e chromosome 2, complete... 676 0.0
CP004380.1 Burkholderia pseudomallei 1026b chromosome 2, complet... 676 0.0
CP010974.1 Burkholderia pseudomallei strain vgh07 chromosome 2, ... 676 0.0
CP009127.1 Burkholderia pseudomallei strain BSR chromosome 2, co... 676 0.0
CP008893.1 Burkholderia pseudomallei HBPUB10303a chromosome 2, c... 676 0.0
CP008912.1 Burkholderia pseudomallei HBPUB10134a chromosome 2, c... 676 0.0
CP008891.1 Burkholderia pseudomallei MSHR5858 chromosome 2, comp... 676 0.0
CP008835.1 Burkholderia pseudomallei strain BGR chromosome 2, co... 676 0.0
CP008782.1 Burkholderia pseudomallei strain Mahidol-1106a chromo... 676 0.0
CP008759.1 Burkholderia pseudomallei strain 1106a chromosome 2, ... 676 0.0
CP008754.1 Burkholderia pseudomallei strain 9 chromosome 2, comp... 676 0.0
CP004002.1 Burkholderia pseudomallei NCTC 13178 chromosome 2, co... 676 0.0
CP003782.1 Burkholderia pseudomallei BPC006 chromosome II, compl... 676 0.0
CP002834.1 Burkholderia pseudomallei 1026b chromosome 2, complet... 676 0.0
CP000573.1 Burkholderia pseudomallei 1106a chromosome II, comple... 676 0.0
CP009163.1 Burkholderia pseudomallei K42 chromosome 2, complete ... 665 0.0
CP016912.1 Burkholderia pseudomallei strain Burk178-Type2 chromo... 660 0.0
CP016910.1 Burkholderia pseudomallei strain Burk178-Type1 chromo... 660 0.0
CP009160.1 Burkholderia pseudomallei TSV 48 chromosome 2, comple... 660 0.0
CP009152.1 Burkholderia pseudomallei MSHR3965 chromosome 2, comp... 660 0.0
CP008910.1 Burkholderia pseudomallei MSHR5848 chromosome 2, comp... 660 0.0
CP008783.1 Burkholderia pseudomallei MSHR5855 chromosome 2, comp... 660 0.0
CP008778.1 Burkholderia pseudomallei 576 chromosome 2, complete ... 660 0.0
CP009550.1 Burkholderia pseudomallei PB08298010 chromosome II, c... 654 0.0
CP025265.1 Burkholderia pseudomallei strain MSHR1435 chromosome ... 649 0.0
CP018412.1 Burkholderia pseudomallei strain 3000015486 chromosom... 649 0.0
CP018411.1 Burkholderia pseudomallei strain 3000015237 chromosom... 649 0.0
CP018409.1 Burkholderia pseudomallei strain 2013833057 chromosom... 649 0.0
CP018407.1 Burkholderia pseudomallei strain 2013833055 chromosom... 649 0.0
CP018398.1 Burkholderia pseudomallei strain 2013746777 chromosom... 649 0.0
CP018397.1 Burkholderia pseudomallei strain 2013746776 chromosom... 649 0.0
CP018394.1 Burkholderia pseudomallei strain 2011756296 chromosom... 649 0.0
CP018392.1 Burkholderia pseudomallei strain 2011756295 chromosom... 649 0.0
CP018388.1 Burkholderia pseudomallei strain 2010007509 chromosom... 649 0.0
CP018386.1 Burkholderia pseudomallei strain 2008724860 chromosom... 649 0.0
CP018419.1 Burkholderia pseudomallei strain 2002734728 chromosom... 649 0.0
CP018371.1 Burkholderia pseudomallei strain 2002721171 chromosom... 649 0.0
CP018370.1 Burkholderia pseudomallei strain 2002721123 chromosom... 649 0.0
CP018367.1 Burkholderia pseudomallei strain 2002721100 chromosome 2 649 0.0
CP017053.1 Burkholderia pseudomallei strain MSHR3763 chromosome ... 649 0.0
CP017051.1 Burkholderia pseudomallei strain MSHR4083 chromosome ... 649 0.0
CP009484.1 Burkholderia pseudomallei MSHR491 chromosome II, comp... 649 0.0
CP009156.1 Burkholderia pseudomallei strain TSV202 chromosome 2,... 649 0.0
CP009234.1 Burkholderia pseudomallei MSHR62 chromosome 2, comple... 649 0.0
CP009269.1 Burkholderia pseudomallei MSHR2243 chromosome 2 sequence 649 0.0
CP009210.1 Burkholderia pseudomallei strain BDP chromosome 2, co... 649 0.0
CP008763.1 Burkholderia pseudomallei strain MSHR346 chromosome 2... 649 0.0
CP008779.1 Burkholderia pseudomallei strain MSHR1655 chromosome ... 649 0.0
CP004369.1 Burkholderia pseudomallei MSHR520 chromosome 2, compl... 649 0.0
CP004043.1 Burkholderia pseudomallei MSHR146 chromosome 2, compl... 649 0.0
CP004024.1 Burkholderia pseudomallei MSHR511 chromosome 2, compl... 649 0.0
CP004004.1 Burkholderia pseudomallei NAU20B-16 chromosome 2, com... 649 0.0
CP006469.1 Burkholderia pseudomallei MSHR305 chromosome 2, compl... 649 0.0
CP009898.1 Burkholderia pseudomallei Pasteur 52237 chromosome 2,... 647 0.0
CP009586.1 Burkholderia pseudomallei strain PHLS 112 chromosome ... 647 0.0
CP008917.1 Burkholderia pseudomallei strain BGK chromosome 2 647 0.0
CP000125.1 Burkholderia pseudomallei 1710b chromosome II, comple... 647 0.0
CP009477.1 Burkholderia pseudomallei MSHR2543 chromosome II, com... 643 1e-180
CP022216.1 Burkholderia thailandensis strain FDAARGOS_242 chromo... 525 4e-145
CP022215.1 Burkholderia thailandensis strain FDAARGOS_241 chromo... 525 4e-145
CP020389.1 Burkholderia thailandensis strain FDAARGOS_237 chromo... 525 4e-145
CP013408.1 Burkholderia thailandensis strain MSMB59 chromosome 2... 525 4e-145
CP009602.1 Burkholderia thailandensis 2002721643 chromosome II, ... 525 4e-145
CP004382.1 Burkholderia thailandensis E254 chromosome 2, complet... 525 4e-145
CP004386.1 Burkholderia thailandensis MSMB59 chromosome 2, compl... 525 4e-145
CP008785.1 Burkholderia thailandensis E264 chromosome 1, complet... 525 4e-145
CP004384.1 Burkholderia thailandensis USAMRU Malaysia #20 chromo... 525 4e-145
CP004118.1 Burkholderia thailandensis E444 chromosome 2, complet... 525 4e-145
CP004098.1 Burkholderia thailandensis 2002721723 chromosome 2, c... 525 4e-145
CP000085.1 Burkholderia thailandensis E264 chromosome II, comple... 525 4e-145
CP013413.1 Burkholderia thailandensis strain 2002721643 chromoso... 499 2e-137
CP010018.1 Burkholderia thailandensis 34 chromosome 2, complete ... 499 2e-137
>CP033704.1 Burkholderia pseudomallei strain FDAARGOS_593 chromosome 2, complete
sequence
Length=3157202
Score = 676 bits (366), Expect = 0.0
Identities = 366/366 (100%), Gaps = 0/366 (0%)
Strand=Plus/Plus
Query 1 ATGATCAAGGACGTTCTACGACTTAAATTCGACGGCAGCCTTTCGCACGATCGGATCGCC 60
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 2603781 ATGATCAAGGACGTTCTACGACTTAAATTCGACGGCAGCCTTTCGCACGATCGGATCGCC 2603840
Query 61 ACGTCGCTGGGCATTTCCAAAAGCGTGGTCACGAAGCACGTCGGACCGGCGGGCGCCGCC 120
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 2603841 ACGTCGCTGGGCATTTCCAAAAGCGTGGTCACGAAGCACGTCGGACCGGCGGGCGCCGCC 2603900
Query 121 GGGCTGGACCGGGCAAGCACCTGCGAGATGGACGAGGGCGAGCGCAAGCGGCGGCTACTC 180
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 2603901 GGGCTGGACCGGGCAAGCACCTGCGAGATGGACGAGGGCGAGCGCAAGCGGCGGCTACTC 2603960
Query 181 GGCAAGCCCATGAGACCAGCGACCTACGTCCAGCCCGATTACGGGCGCATCCATCAGGAA 240
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 2603961 GGCAAGCCCATGAGACCAGCGACCTACGTCCAGCCCGATTACGGGCGCATCCATCAGGAA 2604020
Query 241 CTGCGCCGCAAAGGCGTGACGTTGACGCTGCTGTGGGAGGAGTACCAAGTCGAGTTCGCC 300
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 2604021 CTGCGCCGCAAAGGCGTGACGTTGACGCTGCTGTGGGAGGAGTACCAAGTCGAGTTCGCC 2604080
Query 301 GGCCGGCAAACCTACCGCTCTACGCGCAATTCTGCGAGCACTACAAGGCGTTCACAAAGC 360
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Sbjct 2604081 GGCCGGCAAACCTACCGCTCTACGCGCAATTCTGCGAGCACTACAAGGCGTTCACAAAGC 2604140
Query 361 GTCTGA 366
||||||
Sbjct 2604141 GTCTGA 2604146
outform格式
-outfmt
alignment view options:
0 = Pairwise,
1 = Query-anchored showing identities,
2 = Query-anchored no identities,
3 = Flat query-anchored showing identities,
4 = Flat query-anchored no identities,
5 = BLAST XML,
6 = Tabular,
7 = Tabular with comment lines,
8 = Seqalign (Text ASN.1),
9 = Seqalign (Binary ASN.1),
10 = Comma-separated values,
11 = BLAST archive (ASN.1),
12 = Seqalign (JSON),
13 = Multiple-file BLAST JSON,
14 = Multiple-file BLAST XML2,
15 = Single-file BLAST JSON,
16 = Single-file BLAST XML2,
17 = Sequence Alignment/Map (SAM),
18 = Organism Report
像上面那个如果要输出文件
可以
$ blastn -db mybp -query query.fa -outfmt 7 -out query.txt