Ridge和Lasso回归:Python Scikit-Learn的完整指南
本文将通过一个例子来讲述怎么用scikit-learn来学习Ridge回归和Lasso回归,这两种特殊的线性回归模型 。
1.简介
一般线性回归我们用均方误差作为损失函数,也就是最小二乘法,但是为了防止模型的过拟合,我们在建立线性模型的时候经常需要加入正则化项,一般有L1正则化和L2正则化。
线性回归的L1正则化通常称为Lasso回归,线性回归的L2正则化通常称为Ridge回归,它和一般线性回归的区别是在损失函数上增加了一个L2正则化的项。
2.损失函数
Lasso回归的L1正则化的项有一个常数系数α来调节损失函数的均方差项和正则化项的权重,损失函数表达式如下:
J ( θ ) = 1 2 ( X θ − Y ) T ( X θ − Y ) + α ∣ ∣ θ ∣ ∣ 1 J(\mathbf\theta) = \frac{1}{2}(\mathbf{X\theta} - \mathbf{Y})^T(\mathbf{X\theta} - \mathbf{Y}) + \alpha||\theta||_1 J(θ)=21(Xθ−Y)T(Xθ−Y)+α∣∣θ∣∣1
其中α为常数系数,需要进行调优, ∣ ∣ θ ∣ ∣ 1 ||\theta||_1 ∣∣θ∣∣1 是L1正则化,是对前面的参数加了一个惩罚项,如果 θ \theta θ 取值教大,则优化函数受到较大惩罚。
因此,Lasso回归缩小了系数,有助于降低模型复杂度和多重共线性。
Ridge回归的正则化项是L2范数,而Lasso回归的正则化项是L1范数。具体Ridge回归的损失函数表达式如下:
J ( θ ) = 1 2 ( X θ − Y ) T ( X θ − Y ) + 1 2 α ∣ ∣ θ ∣ ∣ 2 2 J(\mathbf\theta) = \frac{1}{2}(\mathbf{X\theta} - \mathbf{Y})^T(\mathbf{X\theta} - \mathbf{Y}) + \frac{1}{2}\alpha||\theta||_2^2 J(θ)=21(Xθ−Y)T(Xθ−Y)+21α∣∣θ∣∣22
其中α为常数系数,需要进行调优, ∣ ∣ θ ∣ ∣ 2 ||\theta||_2 ∣∣θ∣∣2 是L1正则化,是对前面的参数加了一个惩罚项。
Ridge回归在不抛弃任何一个特征的情况下,缩小了回归系数,使得模型相对而言比较的稳定,但和Lasso回归比,这会使得模型的特征留的特别多,模型解释性差。
3. Scikit-Learn Ridge回归实战
使用Boston house data 的数据集进行模型的拟合。
模型拟合
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
import matplotlib
matplotlib.rcParams.update({'font.size': 12})
from sklearn.datasets import load_boston
from sklearn.cross_validation import train_test_split
from sklearn.linear_model import LinearRegression
from sklearn.linear_model import Ridge
#加载数据集
boston=load_boston()
boston_df=pd.DataFrame(boston.data,columns=boston.feature_names)
print(boston_df.info())
boston_df.head(3)
boston_df.describe()
# 增加一个 Price 属性作为目标值
boston_df['Price']=boston.target
#print (boston_df.head(3))
#newX 训练数据的属性值
newX=boston_df.drop('Price',axis=1)
#newY 训练数据的target值
newY=boston_df['Price']
#交叉验证 分割数据集 30% 数据作为测试
X_train,X_test,y_train,y_test=train_test_split(newX,newY,test_size=0.3,random_state=3)
#线性模型
lr = LinearRegression()
lr.fit(X_train, y_train)
# alpha系数越大,惩罚越大
rr = Ridge(alpha=0.01)
rr.fit(X_train, y_train)
# 使用较大的alpha值
rr100 = Ridge(alpha=100)
rr100.fit(X_train, y_train)
train_score=lr.score(X_train, y_train)
test_score=lr.score(X_test, y_test)
Ridge_train_score = rr.score(X_train,y_train)
Ridge_test_score = rr.score(X_test, y_test)
Ridge_train_score100 = rr100.score(X_train,y_train)
Ridge_test_score100 = rr100.score(X_test, y_test)
print ("linear regression train score:", train_score)
print ("linear regression test score:", test_score)
print ("ridge regression train score low alpha:", Ridge_train_score)
print ("ridge regression test score low alpha:", Ridge_test_score)
print ("ridge regression train score high alpha:", Ridge_train_score100)
print ("ridge regression test score high alpha:", Ridge_test_score100)
#图形化展示 rr.coef_--->模型系数
# markers 为显示的图形
# alpha控制透明度,0为完全透明,1为不透明
plt.plot(rr.coef_,alpha=0.7,linestyle='none',marker='+',markersize=5,color='red',label=r'Ridge; $\alpha = 0.01$',zorder=7)
plt.plot(rr100.coef_,alpha=0.5,linestyle='none',marker='d',markersize=6,color='blue',label=r'Ridge; $\alpha = 100$')
plt.plot(lr.coef_,alpha=0.4,linestyle='none',marker='o',markersize=7,color='green',label='Linear Regression')
plt.xlabel('Coefficient Index',fontsize=16)
plt.ylabel('Coefficient Magnitude',fontsize=16)
plt.legend(fontsize=13,loc=4)
plt.show()
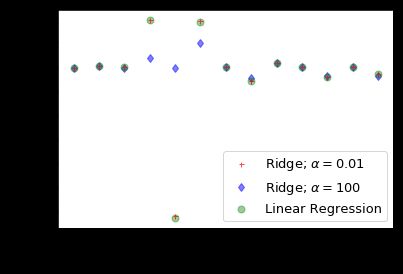
对上图做个简单解释:
- 在X轴中我们使用系数索引(0-12),对于波士顿数据,有13个特征。
- 当α的取比较小的值(0.01),此时模型系数受到较少限制时,系数幅度几乎与线性回归相同。
- 当较大α(100)值,我们看到系数指数为3,4,5的时候,与线性回归情况相比,幅度要小得多。这是使用Ridge回归缩小系数幅度的示例。
超参数α选择
要使用Ridge回归,我们必须要指定超参数 α,你也许有疑问 α 到底改设置成多少?
实际上我们并不知道超参数 α 取多少最好,实际研究是需要在多组自选的α中选择一个最优的。
scikit-learn 提供了一个交叉验证选择最优 α 的 API,可以使用这个API来选择 α 参数,假设我们想在这10个 α 值中选择一个最优的值,代码如下:
from sklearn.linear_model import RidgeCV
ridgecv = RidgeCV(alphas=[0.01, 0.1, 0.5, 1, 5, 7, 10, 30,100, 200])
ridgecv.fit(X_train, y_train)
ridgecv.alpha_ # 0.1
输出结果为:0.1 ,说明在我们给定的这组超参数中, 0.1是最优的 α 值。
4. Scikit-Learn Lasso回归实战
Lasso 回归的正则化项是L1范数,跟 Ridge 回归唯一的区别是不考虑系数的平方,而是考虑幅度。
这种类型的正则化(L1)可以导致零系数,对于输出的评估,一些特征被完全忽略。
因此,Lasso回归不仅有助于减少过度拟合,而且可以帮助我们进行特征选择。
就像Ridge回归一样,正则化参数 α 可以被控制,接下来是使用 sklearn 中的癌症数据集看看效果。不使用上面的波士顿房屋数据,是因为癌症数据集有30个特征属性,而波士顿房屋数据只有13个特征属性。当特征属性比较多的时候,我们可以通过改变正则化参数使用Lasso回归的特征参数的选择。
import math
import matplotlib.pyplot as plt
import pandas as pd
import numpy as np
from sklearn.linear_model import Lasso
from sklearn.linear_model import LinearRegression
from sklearn.datasets import load_breast_cancer
from sklearn.cross_validation import train_test_split
cancer = load_breast_cancer()
#cancer.keys()
cancer_df = pd.DataFrame(cancer.data, columns=cancer.feature_names)
#cancer_df.head(3)
X = cancer.data
Y = cancer.target
X_train,X_test,y_train,y_test=train_test_split(X,Y, test_size=0.3, random_state=31)
lasso = Lasso() # 默认alpha =1
lasso.fit(X_train,y_train)
train_score=lasso.score(X_train,y_train)
test_score=lasso.score(X_test,y_test)
coeff_used = np.sum(lasso.coef_!=0)
print("training score:", train_score )
print ("test score: ", test_score)
print ("number of features used: ", coeff_used)
lasso001 = Lasso(alpha=0.01, max_iter=10e5)
lasso001.fit(X_train,y_train)
train_score001=lasso001.score(X_train,y_train)
test_score001=lasso001.score(X_test,y_test)
coeff_used001 = np.sum(lasso001.coef_!=0)
print ("training score for alpha=0.01:", train_score001 )
print ("test score for alpha =0.01: ", test_score001)
print ("number of features used: for alpha =0.01:", coeff_used001)
lasso00001 = Lasso(alpha=0.0001, max_iter=10e5)
lasso00001.fit(X_train,y_train)
train_score00001=lasso00001.score(X_train,y_train)
test_score00001=lasso00001.score(X_test,y_test)
coeff_used00001 = np.sum(lasso00001.coef_!=0)
print ("training score for alpha=0.0001:", train_score00001 )
print( "test score for alpha =0.0001: ", test_score00001)
print ("number of features used: for alpha =0.0001:", coeff_used00001)
lr = LinearRegression()
lr.fit(X_train,y_train)
lr_train_score=lr.score(X_train,y_train)
lr_test_score=lr.score(X_test,y_test)
print ("LR training score:", lr_train_score )
print ("LR test score: ", lr_test_score)
plt.subplot(1,2,1)
plt.plot(lasso.coef_,alpha=0.7,linestyle='none',marker='*',markersize=5,color='red',label=r'Lasso; $\alpha = 1$',zorder=7) # alpha here is for transparency
plt.plot(lasso001.coef_,alpha=0.5,linestyle='none',marker='d',markersize=6,color='blue',label=r'Lasso; $\alpha = 0.01$') # alpha here is for transparency
plt.xlabel('Coefficient Index',fontsize=16)
plt.ylabel('Coefficient Magnitude',fontsize=16)
plt.legend(fontsize=13,loc=4)
plt.subplot(1,2,2)
plt.plot(lasso.coef_,alpha=0.7,linestyle='none',marker='*',markersize=5,color='red',label=r'Lasso; $\alpha = 1$',zorder=7) # alpha here is for transparency
plt.plot(lasso001.coef_,alpha=0.5,linestyle='none',marker='d',markersize=6,color='blue',label=r'Lasso; $\alpha = 0.01$') # alpha here is for transparency
plt.plot(lasso00001.coef_,alpha=0.8,linestyle='none',marker='v',markersize=6,color='black',label=r'Lasso; $\alpha = 0.00001$') # alpha here is for transparency
plt.plot(lr.coef_,alpha=0.7,linestyle='none',marker='o',markersize=5,color='green',label='Linear Regression',zorder=2)
plt.xlabel('Coefficient Index',fontsize=16)
plt.ylabel('Coefficient Magnitude',fontsize=16)
plt.legend(fontsize=13,loc=4)
plt.tight_layout()
plt.show()
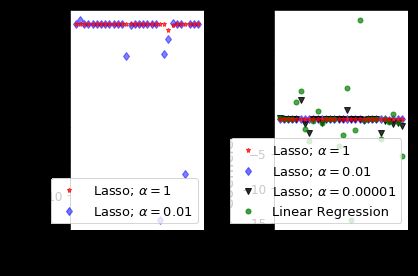
- Lasso 回归中的正则化参数的默认值(alpha)为1;
- 在默认参数的时候,在癌症数据集中的30个特征中,仅使用了4个特征(系数的非零值);
- 训练和测试的分数(仅有4个特征)都很低,得出结论,该默认模型不适合癌症数据集;
- 通过减少alpha和增加迭代次数来减少这种不足,当 alpha = 0.01,非零特征= 10,训练和测试分数增加;
- 首先看左边的一张图,对于α= 1,我们可以看到大多数系数为零或接近零,而 α= 0.01 就多了特征;
- 当我们把 α= 0.0001,非零特征属性数为 22,训练和测试分数与基本线性回归情况相同了;
5. 总结
为了防止模型的过拟合,在建立线性模型的时候经常需要加入(L1、L2)正则化项,也就是对应Lasso和Ridge回归模型,这两种特殊的线性回归模型都有默认的参数以及超参数,在实际运用的过程中一般使用交叉验证选择最优参数,这样可以提高模型的泛化能力。