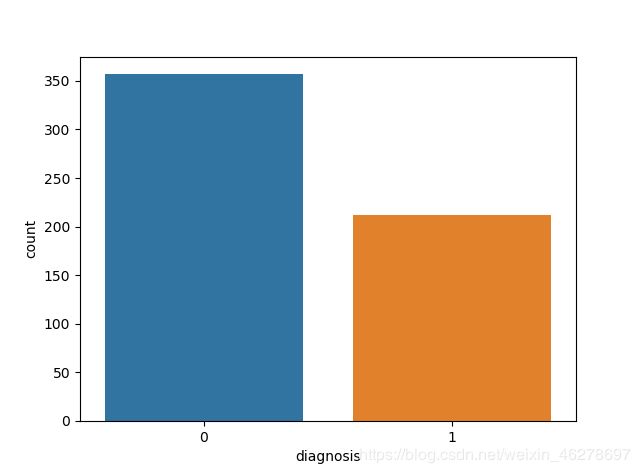
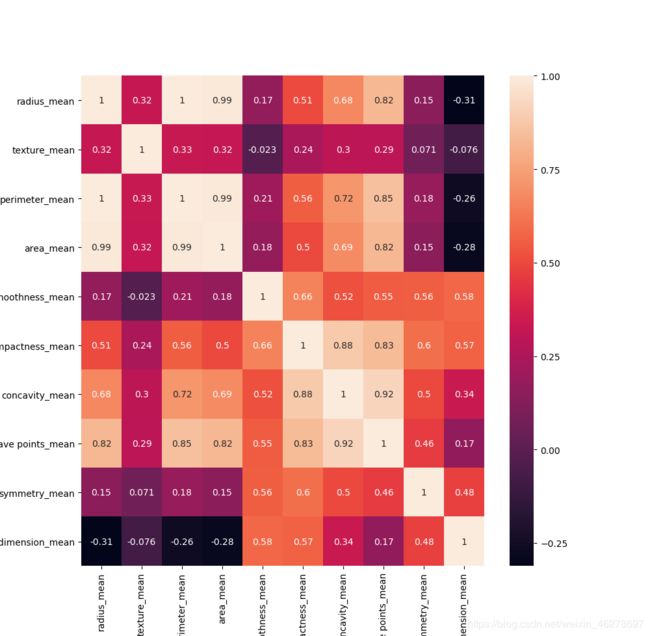
- Mysql的索引失效
不要成为根号三
Mysqlmysql数据库
MySQL的索引失效指的是:尽管在表上建立了索引,但在某些查询场景下,MySQL优化器却没有利用这些索引,从而导致查询走了全表扫描,性能大大降低。下面详细说明几种常见的导致索引失效的情况及其原因:1.对索引列使用函数或表达式问题描述:如果在WHERE子句中对索引列使用函数(如LENGTH(),SUBSTR(),ROUND()等)或进行算术运算(例如id+1=10),MySQL无法直接利用索引,因为
- MySQL 中如何解决深度分页的问题?什么是 MySQL 的主从同步机制?它是如何实现的?如何处理 MySQL 的主从同步延迟?
和道一文字yyds
mysqlandroid数据库
MySQL中如何解决深度分页的问题?在MySQL中,深度分页问题通常是指当使用LIMIT和OFFSET进行分页时,随着分页页码的增加,查询性能显著下降的问题。这是因为MySQL需要扫描OFFSET+LIMIT行,然后丢弃前OFFSET行,导致大量的I/O操作和资源消耗。以下是几种解决深度分页问题的方法:1.游标分页(Cursor-basedPagination)原理:基于有序且唯一的字段(如自增主
- Redis 哈希(Hash)
lsx202406
开发语言
Redis哈希(Hash)概述Redis哈希(Hash)是一种特殊的键值对类型,它允许存储结构化的数据,例如一个对象或记录。每个哈希值可以包含多个字段,每个字段又可以存储一个字符串值。这使得Redis哈希非常适合用于存储对象的属性,如用户信息、配置参数等。哈希数据结构在Redis中,哈希是一个键值对的集合,其中键是字符串,而值也是字符串。哈希中的值可以是以下几种类型:字符串(String)列表(L
- leetcode 922. 按奇偶排序数组 II 简单
圣保罗的大教堂
leetcode每日一题leetcode
给定一个非负整数数组nums,nums中一半整数是奇数,一半整数是偶数。对数组进行排序,以便当nums[i]为奇数时,i也是奇数;当nums[i]为偶数时,i也是偶数。你可以返回任何满足上述条件的数组作为答案。示例1:输入:nums=[4,2,5,7]输出:[4,5,2,7]解释:[4,7,2,5],[2,5,4,7],[2,7,4,5]也会被接受。示例2:输入:nums=[2,3]输出:[2,3
- k8s面试题总结(六)
a_j58
Kubernetes知识点汇总kubernetesjava容器
1.说明一下kubernetes和docker的关系Docker的作用容器镜像管理:Docker可以将应用程序及其依赖打包成一个轻量级的、可移植的容器镜像。容器运行时:Docker提供了一个运行时环境,用于在主机上运行容器。Kubernetes的作用kubernetes是一个容器编排平台,主要用于管理大规模的容器化应用程序。Kubernetes和Docker的关系Docker负责容器的底层运行时:
- 基于GoLang的MMO游戏服务器(三)
帅_shuai_
GOLanggolang服务器游戏go网络
基于GoLang的MMO游戏服务器(三)项目结构Player服务器中每连接一个客户端,相当于一个玩家,封装一个Player结构体typePlayerstruct{Pidint32Connziface.IConnectionXfloat32Yfloat32Zfloat32Vfloat32}初始化一个玩家玩家发送消息给客户端func(p*Player)SendMsg(msgIDuint32,datap
- Redis多线程模型演进
有诺千金
redisredis数据库缓存
一、单线程时代的辉煌(Redis3.x及之前)设计原理:Redis早期采用单Reactor单线程模型,主线程同时处理网络IO和命令执行。这种设计通过事件驱动架构实现高吞吐量,利用epoll/kqueue等系统调用实现非阻塞IO。单线程模型保证了操作的原子性,避免了锁竞争,在内存操作场景下表现出惊人的性能(10万QPS级)。关键特性:串行化命令执行保证原子性内存操作零等待时间规避多线程上下文切换开销
- Redis SCAN 命令详解:安全遍历海量键的利器
有诺千金
redisredis安全数据库
一、SCAN命令的核心价值Redis的KEYS*命令虽然可以遍历所有键,但在生产环境中直接使用可能导致服务阻塞(时间复杂度O(n))。SCAN命令通过游标分批次迭代,实现非阻塞式遍历,成为处理百万级键的安全选择。二、命令语法与参数解析1.基础语法SCANcursor[MATCHpattern][COUNTcount][TYPEtype]2.参数说明参数作用cursor游标值,首次传入0,后续使用前
- 如何让 Git 管理本地项目
有诺千金
gitgit
如何让Git管理本地项目:详细步骤指南Git是最流行的分布式版本控制系统,能够高效管理项目的代码变更历史。以下是将本地项目交给Git管理的完整流程,适用于首次使用Git的开发者。一、前置条件安装Git二、初始化Git仓库进入项目根目录打开终端,使用cd命令切换到你的项目文件夹:cd/path/to/your/project初始化本地仓库执行以下命令,将当前目录变为Git管理的仓库:gitinit这
- 深入解析 synchronized 锁升级:从偏向锁到重量级锁的设计哲学
有诺千金
Java并发编程java
引言在Java并发编程中,synchronized是保证线程安全的核心关键字。但早期的synchronized因直接使用操作系统级互斥锁(MutexLock)而饱受性能诟病。自Java6起,JVM团队引入了锁升级(LockEscalation)机制,通过偏向锁→轻量级锁→重量级锁的渐进式优化,实现了性能与安全的完美平衡。本文将深入剖析每个锁状态的设计思想,揭示其背后的哲学。一、对象头与锁的物理载体
- Java并发编程:深入理解volatile、线程安全陷阱与复合操作
有诺千金
Java并发编程java安全单例模式
一、volatile关键字详解1.核心作用可见性:对volatile变量的写操作立即刷新到主内存,读操作直接读取主内存。有序性:禁止指令重排序(通过内存屏障),确保代码执行顺序符合预期。局限性:不保证原子性(如i++需配合锁或原子类)。2.底层原理JMM层面:插入内存屏障(如StoreLoad屏障),强制缓存同步。硬件层面:依赖CPU的MESI协议实现缓存行失效。3.正确使用场景状态标志:单次写入
- C语言初阶教学----分支和循环(1)
xiaoye_0
c语言c++经验分享职场和发展开发语言
前言对于一个C语言菜鸟来说,说出这些话无疑是大言不惭的,但我有信心也有能力为大家讲好C语言,系统的带大家入门C语言,也希望大家能够相信我,支持我,在接下来的几个月中,我会持续更新这个系列,当然,我的笔记也会同步更新的,感谢大家的支持!如果觉得内容还不错的话,点一个小小的赞和支持吧!课前准备内容总览:分支语句ifswitch循环语句whilefordowhile相信大家都听说过一句话是:C语言是结构
- Linux mkdir 命令
A星空123
linux运维服务器
Linuxmkdir(英文全拼:makedirectory)命令用于创建目录。语法mkdir[-p]dirName参数说明:-p确保目录名称存在,不存在的就建一个。实例在工作目录下,建立一个名为runoob的子目录:mkdirrunoob在工作目录下的runoob2目录中,建立一个名为test的子目录。若runoob2目录原本不存在,则建立一个。(注:本例若不加-p参数,且原本runoob2目录不
- Linux ls 命令
A星空123
linux运维服务器
Linuxls(英文全拼:listdirectorycontents)命令用于显示指定工作目录下之内容(列出目前工作目录所含的文件及子目录)。语法ls[-alrtAFR][name...]参数:-a显示所有文件及目录(.开头的隐藏文件也会列出)-d只列出目录(不递归列出目录内的文件)。-l以长格式显示文件和目录信息,包括权限、所有者、大小、创建时间等。-r倒序显示文件和目录。-t将按照修改时间排序
- Unity程序开发:5.UI系统
风不归Alkaid
Unity游戏程序开发讲解unityui游戏引擎c#
5.UI系统Unity提供了一个强大的UI系统,用于创建和管理游戏中的用户界面。以下是UnityUI系统的详细介绍和示例代码,包括基本UI组件、UI布局以及处理UI事件。基本UI组件TextText组件用于显示文字。示例:创建Text组件usingUnityEngine;usingUnityEngine.UI;publicclassCreateText:MonoBehaviour{voidStar
- 在 Ubuntu20.04 上安装 Docker 并部署 Dify
奕997
ubuntudockerlinux
1.安装Docker1.1更新系统软件包列表在终端执行以下命令更新软件包列表:sudoaptupdate1.2安装必要的依赖包为了能够通过HTTPS安装Docker,我们需要先安装一些必备包:sudoaptinstallapt-transport-httpsca-certificatescurlsoftware-properties-common1.3添加Docker官方GPG密钥执行以下命令,将
- threeJs+vue 添加控制面板gui,修改几何体和页面的背景色
资深前端之路
threeJs前端threeJs
嗨,我是小路。今天主要和大家分享的主题是“threeJs+vue添加控制面板gui,修改几何体和页面的背景色”。在现代Web开发中,创建引人入胜的交互式3D内容已成为提升用户体验的关键因素之一。而将Three.js与Vue结合使用,可以让你轻松构建出既美观又功能强大的3D应用程序。今天,我们将介绍如何通过添加一个直观的GUI控制面板来进一步增强你的Three.js项目,使用户能够实时调整几何体的颜
- 如何测试国内地区网站的访问情况?
资深前端之路
学习学习
当自己服务器不稳定时,可以通过以下的方法检测到国内哪些区域访问的速度,知道哪个地区的网络好,哪个区域网络较差。1、收集可以进行网站多端测试的接口。如www.ping.cn2、将自己想要测试的域名,填入输入框,不需要带入http或者https。如百度:www.baidu.com,检测结果可以在下图中,显示整体的访问情况;下面的检测结果也有详细的列表数据。
- ajax检测超时,jquery – 使用AJAX时检查会话超时
是TuTu兔
ajax检测超时
我有一个ColdFusion页面,用户可以打开一个模态并查看有关一行数据的更多信息.但是,如果用户在页面上的时间超过默认的20分钟会话超时,则会抛出错误,因为它正在查找会话变量但无法找到它们.我理解如何使用服务器端代码捕获这个,但我似乎无法通过AJAX调用来成功确定会话是否仍然存在.这是当用户点击按钮打开模态时触发的AJAX代码.基本上它正在检查会话是否与CFC中的函数一起存在.我的问题是,它总是
- ubantu(20.04)+ollama+dify+摩尔线程S80,新手避坑指南
chaonghoeoh
ubuntudocker
前言,ubantu(20.04)+ollama+dify+摩尔线程S80,在组装过程中,作为新手会遇到不少的坑,特别写了这系列教程,是为增强新手上路的填坑能力。同时,摩尔线程作为养成系列的显卡,在运用场景上还是比较少,希望更多感兴趣的人一起探讨。一、ubantu(20.04)常遇到的问题第一个,下载安装好后,第一个先设置后语言支持和地区格式,要是安装的第一步选择好语言会更好,新手往往会忽视。第二个
- Unity3D-UI--Layout组件
superlinmeng
Unity3Dunity
Layout组件自动排版LayoutGroupVerticalLayoutGroup垂直布局【垂直布局组】组件将其子布局元素彼此重叠。它们的高度由各自的最小高度,首选高度和柔性高度决定,具体取决于以下模型:VerticalLayoutGroup表格布局ContentSizeFitter宽高适配组件AspectRatioFitter根据比例控制当前的宽高AspectMode:控制的模式:Layout
- 使用FastAPI进行可视化部署
AllYoung_362
AIGC人工智能深度学习算法llamachatgpt
文章目录一、FastAPI介绍二、环境配置三、示例代码1.app.py代码如下2.websocket_handler.py代码如下3.运行app4.遇到的问题与解决一、FastAPI介绍FastAPI是一个高性能的PythonWeb框架,它基于Starlette并利用了Python类型提示的优势。它可以帮助我们快速构建具有强大功能的Web应用程序。二、环境配置依赖库介绍:Uvicorn是一个基于a
- 数据结构漫谈
你一身傲骨怎能输
数据结构数据结构
数据结构是计算机科学中一种组织和存储数据的方式,它使得数据可以高效地被访问和修改。数据结构可以分为线性数据结构和非线性数据结构两大类,以下是一些常见的数据结构:线性数据结构数组(Array)一组连续的内存空间,用来存储相同类型的数据。支持快速的随机访问,但插入和删除操作可能较慢。链表(LinkedList)由一系列节点组成,每个节点包含数据和指向下一个节点的指针。插入和删除操作相对较快,但不支持快
- 【Elasticsearch】一文读懂ES向量搜索:原理剖析与技术全景
程序员大任
ElasticSearchelasticsearch
注:本文若未说明ES版本则为7.10,其他版本会特别标记,由于ES版本不同,部分差异较大,具体请以官方文档为准一、向量搜索的核心原理1.1向量化表示的本质现代AI技术将文本、图像等非结构化数据转化为高维向量(通常128-1024维),这些向量在数学空间中携带语义特征。如:文本嵌入(Embedding):BERT等模型生成768维向量图像特征:ResNet提取2048维特征向量1.2向量搜索简介向量
- 使用 malloc 函数创建和操作二维整型数组
共享家9527
c++c语言算法数据结构
目录一、引言二、代码实现三、代码详解(一)头文件引入(二)定义数组维度(三)动态分配二维数组内存(四)初始化二维数组(五)输出二维数组(六)释放内存四、总结一、引言在C语言编程中,动态内存分配是一项非常重要的技能。它允许我们在程序运行时根据实际需求分配内存,提高内存使用效率。malloc函数是C语言中用于动态内存分配的重要工具之一。本文将详细介绍如何使用malloc函数模拟开辟一个3*5的整型二维
- 每日一题之k倍区间
Ace'
算法数据结构
题目描述给定一个长度为N的数列,A1,A2,⋯AN,如果其中一段连续的子序列Ai,Ai+1,⋯Aj(i≤j)之和是K的倍数,我们就称这个区间[i,j]是K倍区间。你能求出数列中总共有多少个K倍区间吗?输入描述第一行包含两个整数N和K(1≤N,K≤105)。以下N行每行包含一个整数Ai(1≤Ai≤105)输出描述输出一个整数,代表K倍区间的数目。#includeusingnamespacestd;i
- 动态会话管理:保持用户活跃状态的实践
t0_54coder
Java语言开发教程apachetomcatssl个人开发
动态会话管理:保持用户活跃状态的实践在Web应用开发中,会话管理是一个重要的环节。用户在进行长时间的操作时,我们希望他们的会话保持活跃,而不是因为服务器端的会话超时而被迫重新登录。本文将通过一个实际的JavaWeb应用示例,展示如何动态地管理用户会话,以保持用户在客户端活跃时会话不超时,并在用户空闲一定时间后自动登出。问题背景在之前的示例中,我们讨论了如何在会话过期时重定向到登录页面。但这种方法存
- 基于YOLOv5的无人超市商品检测:食品、饮料、零食与家居用品
深度学习&目标检测实战项目
YOLO目标跟踪深度学习人工智能ui
引言随着人工智能技术的快速发展,尤其是计算机视觉的提升,无人超市的概念逐渐成为现实。在无人超市中,商品的智能化管理和检测是其顺利运行的关键。商品检测不仅要实现高效、准确的物品识别,还要支持多种商品类别的实时检测,以保证购物体验的顺畅与安全。在此背景下,深度学习与目标检测算法,如YOLOv5,成为了实现这一目标的重要工具。YOLOv5作为目前最先进且高效的目标检测算法之一,其应用范围广泛,包括人脸检
- AI驱动的前端技术选型:告别选择困难症,拥抱高效开发时代
liangruimi
人工智能前端
在快节奏的互联网时代,选择合适的前端技术栈对于项目的成功至关重要。然而,面对层出不穷的新技术和框架,前端开发者常常面临着技术选型难题,例如技术更新迭代快,选择困难,以及传统方法效率低下等问题。因此,利用AI代码生成器等AI工具辅助前端技术选型,已成为提升效率、降低风险的关键。本文将探讨如何借助AI的力量,轻松应对前端技术选型挑战,高效完成项目开发。AI驱动的前端技术选型:现状与挑战目前,前端技术栈
- Qt-DAB 开源项目指南
芮逸炯Conqueror
Qt-DAB开源项目指南qt-dabQt-DAB,ageneralsoftwareDAB(DAB+)decoderwitha(slight)focusonshowingthesignal项目地址:https://gitcode.com/gh_mirrors/qt/qt-dab项目介绍Qt-DAB是一个基于Qt框架的数字音频广播(DAB)解决方案。该项目旨在提供一个用于接收、解码和播放DAB广播信号
- Java 并发包之线程池和原子计数
lijingyao8206
Java计数ThreadPool并发包java线程池
对于大数据量关联的业务处理逻辑,比较直接的想法就是用JDK提供的并发包去解决多线程情况下的业务数据处理。线程池可以提供很好的管理线程的方式,并且可以提高线程利用率,并发包中的原子计数在多线程的情况下可以让我们避免去写一些同步代码。
这里就先把jdk并发包中的线程池处理器ThreadPoolExecutor 以原子计数类AomicInteger 和倒数计时锁C
- java编程思想 抽象类和接口
百合不是茶
java抽象类接口
接口c++对接口和内部类只有简介的支持,但在java中有队这些类的直接支持
1 ,抽象类 : 如果一个类包含一个或多个抽象方法,该类必须限定为抽象类(否者编译器报错)
抽象方法 : 在方法中仅有声明而没有方法体
package com.wj.Interface;
- [房地产与大数据]房地产数据挖掘系统
comsci
数据挖掘
随着一个关键核心技术的突破,我们已经是独立自主的开发某些先进模块,但是要完全实现,还需要一定的时间...
所以,除了代码工作以外,我们还需要关心一下非技术领域的事件..比如说房地产
&nb
- 数组队列总结
沐刃青蛟
数组队列
数组队列是一种大小可以改变,类型没有定死的类似数组的工具。不过与数组相比,它更具有灵活性。因为它不但不用担心越界问题,而且因为泛型(类似c++中模板的东西)的存在而支持各种类型。
以下是数组队列的功能实现代码:
import List.Student;
public class
- Oracle存储过程无法编译的解决方法
IT独行者
oracle存储过程
今天同事修改Oracle存储过程又导致2个过程无法被编译,流程规范上的东西,Dave 这里不多说,看看怎么解决问题。
1. 查看无效对象
XEZF@xezf(qs-xezf-db1)> select object_name,object_type,status from all_objects where status='IN
- 重装系统之后oracle恢复
文强chu
oracle
前几天正在使用电脑,没有暂停oracle的各种服务。
突然win8.1系统奔溃,无法修复,开机时系统 提示正在搜集错误信息,然后再开机,再提示的无限循环中。
无耐我拿出系统u盘 准备重装系统,没想到竟然无法从u盘引导成功。
晚上到外面早了一家修电脑店,让人家给装了个系统,并且那哥们在我没反应过来的时候,
直接把我的c盘给格式化了 并且清理了注册表,再装系统。
然后的结果就是我的oracl
- python学习二( 一些基础语法)
小桔子
pthon基础语法
紧接着把!昨天没看继续看django 官方教程,学了下python的基本语法 与c类语言还是有些小差别:
1.ptyhon的源文件以UTF-8编码格式
2.
/ 除 结果浮点型
// 除 结果整形
% 除 取余数
* 乘
** 乘方 eg 5**2 结果是5的2次方25
_&
- svn 常用命令
aichenglong
SVN版本回退
1 svn回退版本
1)在window中选择log,根据想要回退的内容,选择revert this version或revert chanages from this version
两者的区别:
revert this version:表示回退到当前版本(该版本后的版本全部作废)
revert chanages from this versio
- 某小公司面试归来
alafqq
面试
先填单子,还要写笔试题,我以时间为急,拒绝了它。。时间宝贵。
老拿这些对付毕业生的东东来吓唬我。。
面试官很刁难,问了几个问题,记录下;
1,包的范围。。。public,private,protect. --悲剧了
2,hashcode方法和equals方法的区别。谁覆盖谁.结果,他说我说反了。
3,最恶心的一道题,抽象类继承抽象类吗?(察,一般它都是被继承的啊)
4,stru
- 动态数组的存储速度比较 集合框架
百合不是茶
集合框架
集合框架:
自定义数据结构(增删改查等)
package 数组;
/**
* 创建动态数组
* @author 百合
*
*/
public class ArrayDemo{
//定义一个数组来存放数据
String[] src = new String[0];
/**
* 增加元素加入容器
* @param s要加入容器
- 用JS实现一个JS对象,对象里有两个属性一个方法
bijian1013
js对象
<html>
<head>
</head>
<body>
用js代码实现一个js对象,对象里有两个属性,一个方法
</body>
<script>
var obj={a:'1234567',b:'bbbbbbbbbb',c:function(x){
- 探索JUnit4扩展:使用Rule
bijian1013
java单元测试JUnitRule
在上一篇文章中,讨论了使用Runner扩展JUnit4的方式,即直接修改Test Runner的实现(BlockJUnit4ClassRunner)。但这种方法显然不便于灵活地添加或删除扩展功能。下面将使用JUnit4.7才开始引入的扩展方式——Rule来实现相同的扩展功能。
1. Rule
&n
- [Gson一]非泛型POJO对象的反序列化
bit1129
POJO
当要将JSON数据串反序列化自身为非泛型的POJO时,使用Gson.fromJson(String, Class)方法。自身为非泛型的POJO的包括两种:
1. POJO对象不包含任何泛型的字段
2. POJO对象包含泛型字段,例如泛型集合或者泛型类
Data类 a.不是泛型类, b.Data中的集合List和Map都是泛型的 c.Data中不包含其它的POJO
- 【Kakfa五】Kafka Producer和Consumer基本使用
bit1129
kafka
0.Kafka服务器的配置
一个Broker,
一个Topic
Topic中只有一个Partition() 1. Producer:
package kafka.examples.producers;
import kafka.producer.KeyedMessage;
import kafka.javaapi.producer.Producer;
impor
- lsyncd实时同步搭建指南——取代rsync+inotify
ronin47
1. 几大实时同步工具比较 1.1 inotify + rsync
最近一直在寻求生产服务服务器上的同步替代方案,原先使用的是 inotify + rsync,但随着文件数量的增大到100W+,目录下的文件列表就达20M,在网络状况不佳或者限速的情况下,变更的文件可能10来个才几M,却因此要发送的文件列表就达20M,严重减低的带宽的使用效率以及同步效率;更为要紧的是,加入inotify
- java-9. 判断整数序列是不是二元查找树的后序遍历结果
bylijinnan
java
public class IsBinTreePostTraverse{
static boolean isBSTPostOrder(int[] a){
if(a==null){
return false;
}
/*1.只有一个结点时,肯定是查找树
*2.只有两个结点时,肯定是查找树。例如{5,6}对应的BST是 6 {6,5}对应的BST是
- MySQL的sum函数返回的类型
bylijinnan
javaspringsqlmysqljdbc
今天项目切换数据库时,出错
访问数据库的代码大概是这样:
String sql = "select sum(number) as sumNumberOfOneDay from tableName";
List<Map> rows = getJdbcTemplate().queryForList(sql);
for (Map row : rows
- java设计模式之单例模式
chicony
java设计模式
在阎宏博士的《JAVA与模式》一书中开头是这样描述单例模式的:
作为对象的创建模式,单例模式确保某一个类只有一个实例,而且自行实例化并向整个系统提供这个实例。这个类称为单例类。 单例模式的结构
单例模式的特点:
单例类只能有一个实例。
单例类必须自己创建自己的唯一实例。
单例类必须给所有其他对象提供这一实例。
饿汉式单例类
publ
- javascript取当月最后一天
ctrain
JavaScript
<!--javascript取当月最后一天-->
<script language=javascript>
var current = new Date();
var year = current.getYear();
var month = current.getMonth();
showMonthLastDay(year, mont
- linux tune2fs命令详解
daizj
linuxtune2fs查看系统文件块信息
一.简介:
tune2fs是调整和查看ext2/ext3文件系统的文件系统参数,Windows下面如果出现意外断电死机情况,下次开机一般都会出现系统自检。Linux系统下面也有文件系统自检,而且是可以通过tune2fs命令,自行定义自检周期及方式。
二.用法:
Usage: tune2fs [-c max_mounts_count] [-e errors_behavior] [-g grou
- 做有中国特色的程序员
dcj3sjt126com
程序员
从出版业说起 网络作品排到靠前的,都不会太难看,一般人不爱看某部作品也是因为不喜欢这个类型,而此人也不会全不喜欢这些网络作品。究其原因,是因为网络作品都是让人先白看的,看的好了才出了头。而纸质作品就不一定了,排行榜靠前的,有好作品,也有垃圾。 许多大牛都是写了博客,后来出了书。这些书也都不次,可能有人让为不好,是因为技术书不像小说,小说在读故事,技术书是在学知识或温习知识,有
- Android:TextView属性大全
dcj3sjt126com
textview
android:autoLink 设置是否当文本为URL链接/email/电话号码/map时,文本显示为可点击的链接。可选值(none/web/email/phone/map/all) android:autoText 如果设置,将自动执行输入值的拼写纠正。此处无效果,在显示输入法并输
- tomcat虚拟目录安装及其配置
eksliang
tomcat配置说明tomca部署web应用tomcat虚拟目录安装
转载请出自出处:http://eksliang.iteye.com/blog/2097184
1.-------------------------------------------tomcat 目录结构
config:存放tomcat的配置文件
temp :存放tomcat跑起来后存放临时文件用的
work : 当第一次访问应用中的jsp
- 浅谈:APP有哪些常被黑客利用的安全漏洞
gg163
APP
首先,说到APP的安全漏洞,身为程序猿的大家应该不陌生;如果抛开安卓自身开源的问题的话,其主要产生的原因就是开发过程中疏忽或者代码不严谨引起的。但这些责任也不能怪在程序猿头上,有时会因为BOSS时间催得紧等很多可观原因。由国内移动应用安全检测团队爱内测(ineice.com)的CTO给我们浅谈关于Android 系统的开源设计以及生态环境。
1. 应用反编译漏洞:APK 包非常容易被反编译成可读
- C#根据网址生成静态页面
hvt
Web.netC#asp.nethovertree
HoverTree开源项目中HoverTreeWeb.HVTPanel的Index.aspx文件是后台管理的首页。包含生成留言板首页,以及显示用户名,退出等功能。根据网址生成页面的方法:
bool CreateHtmlFile(string url, string path)
{
//http://keleyi.com/a/bjae/3d10wfax.htm
stri
- SVG 教程 (一)
天梯梦
svg
SVG 简介
SVG 是使用 XML 来描述二维图形和绘图程序的语言。 学习之前应具备的基础知识:
继续学习之前,你应该对以下内容有基本的了解:
HTML
XML 基础
如果希望首先学习这些内容,请在本站的首页选择相应的教程。 什么是SVG?
SVG 指可伸缩矢量图形 (Scalable Vector Graphics)
SVG 用来定义用于网络的基于矢量
- 一个简单的java栈
luyulong
java数据结构栈
public class MyStack {
private long[] arr;
private int top;
public MyStack() {
arr = new long[10];
top = -1;
}
public MyStack(int maxsize) {
arr = new long[maxsize];
top
- 基础数据结构和算法八:Binary search
sunwinner
AlgorithmBinary search
Binary search needs an ordered array so that it can use array indexing to dramatically reduce the number of compares required for each search, using the classic and venerable binary search algori
- 12个C语言面试题,涉及指针、进程、运算、结构体、函数、内存,看看你能做出几个!
刘星宇
c面试
12个C语言面试题,涉及指针、进程、运算、结构体、函数、内存,看看你能做出几个!
1.gets()函数
问:请找出下面代码里的问题:
#include<stdio.h>
int main(void)
{
char buff[10];
memset(buff,0,sizeof(buff));
- ITeye 7月技术图书有奖试读获奖名单公布
ITeye管理员
活动ITeye试读
ITeye携手人民邮电出版社图灵教育共同举办的7月技术图书有奖试读活动已圆满结束,非常感谢广大用户对本次活动的关注与参与。
7月试读活动回顾:
http://webmaster.iteye.com/blog/2092746
本次技术图书试读活动的优秀奖获奖名单及相应作品如下(优秀文章有很多,但名额有限,没获奖并不代表不优秀):
《Java性能优化权威指南》