李宏毅 机器学习 作业 2 Hungyi.Li Machine Learning HW2 逻辑回归法实现以及优化
关于“一个属性有几个固定选项,怎么表达”的问题,数据采用one-hot,每个选项独立成一个属性,loss用交叉熵计算,这样更科学。这里不用过多操心,数据给出已经处理好了。
有一段可能是助教给的参考代码,不过参考代码有些漏洞(在代码下方贴出),需要修改下,然后还有一定的调参提升空间。
参考代码如下:
import os, sys
import numpy as np
from random import shuffle
import argparse
from math import log, floor
import pandas as pd
# If you wish to get the same shuffle result
# np.random.seed(2401)
def load_data(train_data_path, train_label_path, test_data_path):
X_train = pd.read_csv(train_data_path, sep=',', header=0)
X_train = np.array(X_train.values)
Y_train = pd.read_csv(train_label_path, sep=',', header=0)
Y_train = np.array(Y_train.values)
X_test = pd.read_csv(test_data_path, sep=',', header=0)
X_test = np.array(X_test.values)
return (X_train, Y_train, X_test)
def _shuffle(X, Y):
randomize = np.arange(len(X))
np.random.shuffle(randomize)
return (X[randomize], Y[randomize])
def normalize(X_all, X_test):
# Feature normalization with train and test X
X_train_test = np.concatenate((X_all, X_test))
mu = (sum(X_train_test) / X_train_test.shape[0])
sigma = np.std(X_train_test, axis=0)
mu = np.tile(mu, (X_train_test.shape[0], 1))
sigma = np.tile(sigma, (X_train_test.shape[0], 1))
X_train_test_normed = (X_train_test - mu) / sigma
# Split to train, test again
X_all = X_train_test_normed[0:X_all.shape[0]]
X_test = X_train_test_normed[X_all.shape[0]:]
return X_all, X_test
def split_valid_set(X_all, Y_all, percentage):
all_data_size = len(X_all)
valid_data_size = int(floor(all_data_size * percentage))#这个比例反了。
X_all, Y_all = _shuffle(X_all, Y_all)
X_train, Y_train = X_all[0:valid_data_size], Y_all[0:valid_data_size]
X_valid, Y_valid = X_all[valid_data_size:], Y_all[valid_data_size:]
return X_train, Y_train, X_valid, Y_valid
def sigmoid(z):
res = 1 / (1.0 + np.exp(-z))
return np.clip(res, 1e-8, 1-(1e-8))
def valid(w, b, X_valid, Y_valid):
valid_data_size = len(X_valid)
z = (np.dot(X_valid, np.transpose(w)) + b)
y = sigmoid(z)
y_ = np.around(y)
result = (np.squeeze(Y_valid) == y_)
print('Validation acc = %f' % (float(result.sum()) / valid_data_size))
return
def train(X_all, Y_all, save_dir):
# Split a 10%-validation set from the training set
valid_set_percentage = 0.1
X_train, Y_train, X_valid, Y_valid = split_valid_set(X_all, Y_all, valid_set_percentage)
# Initiallize parameter, hyperparameter
w = np.zeros((106,))
b = np.zeros((1,))
l_rate = 0.1
batch_size = 32
train_data_size = len(X_train)
step_num = int(floor(train_data_size / batch_size))
epoch_num = 1000
save_param_iter = 50
# Start training
total_loss = 0.0
for epoch in range(1, epoch_num):
# Do validation and parameter saving
if (epoch) % save_param_iter == 0:
print('=====Saving Param at epoch %d=====' % epoch)
if not os.path.exists(save_dir):
os.mkdir(save_dir)
np.savetxt(os.path.join(save_dir, 'w'), w)
np.savetxt(os.path.join(save_dir, 'b'), [b,])
print('epoch avg loss = %f' % (total_loss / (float(save_param_iter) * train_data_size)))
total_loss = 0.0
valid(w, b, X_valid, Y_valid)
# Random shuffle
X_train, Y_train = _shuffle(X_train, Y_train)
# Train with batch
for idx in range(step_num):
X = X_train[idx*batch_size:(idx+1)*batch_size]
Y = Y_train[idx*batch_size:(idx+1)*batch_size]
z = np.dot(X, np.transpose(w)) + b
y = sigmoid(z)
cross_entropy = -1 * (np.dot(np.squeeze(Y), np.log(y)) + np.dot((1 - np.squeeze(Y)), np.log(1 - y)))
total_loss += cross_entropy
w_grad = np.sum(-1 * X * (np.squeeze(Y) - y).reshape((batch_size,1)), axis=0)
b_grad = np.sum(-1 * (np.squeeze(Y) - y))
# SGD updating parameters
w = w - l_rate * w_grad
b = b - l_rate * b_grad
return
def infer(X_test, save_dir, output_dir):
test_data_size = len(X_test)
# Load parameters
print('=====Loading Param from %s=====' % save_dir)
w = np.loadtxt(os.path.join(save_dir, 'w'))
b = np.loadtxt(os.path.join(save_dir, 'b'))
# predict
z = (np.dot(X_test, np.transpose(w)) + b)
y = sigmoid(z)
y_ = np.around(y)
print('=====Write output to %s =====' % output_dir)
if not os.path.exists(output_dir):
os.mkdir(output_dir)
output_path = os.path.join(output_dir, 'log_prediction.csv')
with open(output_path, 'w') as f:
f.write('id,label\n')
for i, v in enumerate(y_):
f.write('%d,%d\n' %(i+1, v))
return
def main(opts):
# Load feature and label
X_all, Y_all, X_test = load_data(opts.train_data_path, opts.train_label_path, opts.test_data_path)
# Normalization
X_all, X_test = normalize(X_all, X_test)
# To train or to infer
if opts.train:
train(X_all, Y_all, opts.save_dir)
elif opts.infer:
infer(X_test, opts.save_dir, opts.output_dir)
else:
print("Error: Argument --train or --infer not found")
return
if __name__ == '__main__':
parser = argparse.ArgumentParser(description='Logistic Regression with Gradient Descent Method')
group = parser.add_mutually_exclusive_group()
group.add_argument('--train', action='store_true', default=False,
dest='train', help='Input --train to Train')
group.add_argument('--infer', action='store_true',default=False,
dest='infer', help='Input --infer to Infer')
parser.add_argument('--train_data_path', type=str,
default='feature/X_train', dest='train_data_path',
help='Path to training data')
parser.add_argument('--train_label_path', type=str,
default='feature/Y_train', dest='train_label_path',
help='Path to training data\'s label')
parser.add_argument('--test_data_path', type=str,
default='feature/X_test', dest='test_data_path',
help='Path to testing data')
parser.add_argument('--save_dir', type=str,
default='logistic_params/', dest='save_dir',
help='Path to save the model parameters')
parser.add_argument('--output_dir', type=str,
default='logistic_output/', dest='output_dir',
help='Path to save the model parameters')
opts = parser.parse_args()
main(opts)代码错误有几点:
1.每一个epoch中,用于计算cross_entropy从而统计进total_loss的数据量,是step_num*batch_size,小于train_data_size。而ta
它用的是train_data_size。
这两个数据对比:结果证实,确实有点偏差,不过因为他只是打印,不是训练过程,所以看起来也没影响结果。
2.split_valid_set中,训练集和验证集划分错了,用验证集的比例,提取了训练集。
这个也很明显,会对结果产生显著影响
3.感谢网友weixin_42183288提醒,不过此处确实没错:交叉熵隐含了sum操作!
因为np.dot是点积,点积已经等于sum了,batch_size个样本,最终只输出一维结果,对于batch_size个样本,loss既然是求和,grad也应该是求和,grad要和loss保持同步,所以grad就是sum。
print('epoch avg loss = %f' % (total_loss / (float(save_param_iter) * train_data_size)))从这句打印也看得出,loss确实是所有样本的和,然后才除以迭代epoch次数和data_size,平均到每个样本。
但是打印归打印,这句不是也说了是avg loss嘛!实际的total loss,还是针对batch的sum。
其实这根本不是应该纠结的点!!!这可能是接触代码不多的新手都会碰到的疑惑:
“课程中是除以m,也就是mean”
“loss要用sum还是mean?”
“grad要用sum还是mean?”
“为什么我用mean比用sum感觉要更准一点?”
本质上,这是没区别的,梯度下降,下降的是什么?loss的值!具体loss或者w的大小,只与计算机对小数的表达能力有关(这个角度看,mean反而不如sum准),而不太可能上升到模型准确度差异的高度!教科书是为了示意,如果他想精准地表达梯度,当然要除以m,可是实际计算当中,区别不大。
无论你的grad是用了sum还是mean,最终都是一个数值(乘以参数数量),是数值,在梯度下降时就逃不过乘以l_rate这一环,如果你觉得你用了mean比sum训练结果更准,可以把sum版本对应的l_rate调小一点(实际训练中往往是变动的学习率,越到后期越小)
w_grad = np.mean(-1 * X * (np.squeeze(Y) - y).reshape((batch_size,1)), axis=0)
b_grad = np.mean(-1 * (np.squeeze(Y) - y))
# SGD updating parameters
w = w - l_rate * w_grad
b = b - l_rate * b_grad
4.还有个什么错误我忘了。。。。
上传kaggle,这是两条分数分段线:
0.84952高分
0.84215及格
验证集分配错误的,也够及格
0.84803
优化的分配比例的:
X_valid,Y_valid = X_all[0:valid_data_size],Y_all[0:valid_data_size]
X_train,Y_train = X_all[valid_data_size:],Y_all[valid_data_size:]
0.85159
下边是调整其他参数,batch_size和epoch等:
batch_size提升到64,epoch到5000
0.85356
batch_size提升到64,epoch到5000,l_rate到0.5
0.84201
会下降,猜测可能是没用adagrad,这里不收敛了。
batch_size提升到64,epoch到5000,l_rate改到0.2,
0.85036
比0.5要强,不过还是比0.1差些,也许有运气成分,每次训练也不一样。
batch_size提升到64,epoch到5000,l_rate改到0.05,
0.85380目前最优,可能是epoch数量比较大,抵消了learning rate的降低效应。可以明显看到,learning调大明显有收敛问题,所以adagrad可能效果更好。
还有训练,集和验证集的比例可以调:
batch_size提升到64,epoch到5000,l_rate改到0.05,验证集改到0.3(默认0.1)
validation acc = 0.851761
实际分数0.85417目前最优,可能说明之前的有过拟合现象。
其他不动,验证集比例改回0.1重新测
validation acc = 0.860258
分数:0.85417,居然一样
本地validation acc更高一些,实际分数还一样,是有一点过拟合吧??!!
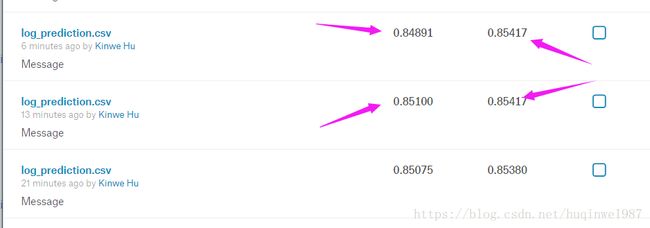
调了一下记录,可以看到倒数第二次,虽然公开的也是0.85417,和最后一次一样,但是最后一次在private衰减到了0.84891,证明最后一次确实比倒数第二次过拟合。
但是过拟合和过拟合也不一样,有两种过拟合,这里说明最后一次是针对线上公开成绩的过拟合,private set发生衰减。
前边说的是针对验证集的过拟合,线上答案发生衰减。
紫色字体不算绝对严谨,因为需要更细致的测试对比,每次训练本来就有微小的差距。这里主要讨论可能存在的问题和优化思路,因为时间关系和运算量的关系,就不在这纠结了,先结了。
实际跑了一下adagrad,效果飘忽,调起来比较麻烦,时间问题,略过
w_s_gra = np.zeros(106)
b_s_gra = np.zeros(1)
for idx in range(step_num):
X = X_train[idx*batch_size:(idx+1)*batch_size]
Y = Y_train[idx*batch_size:(idx+1)*batch_size]
z = np.dot(X,np.transpose(w))+b
y = sigmoid(z)
cross_entropy = -1*(np.dot(np.squeeze(Y),np.log(y))+np.dot(1-np.squeeze(Y),np.log(1-y)))
total_loss += cross_entropy
w_grad = np.mean(-1*X*(np.squeeze(Y)-y).reshape((batch_size,1)),axis=0)
b_grad = np.mean(-1*(np.squeeze(Y)-y))
w_s_gra += w_grad**2
b_s_gra += b_grad**2
w_ada = np.sqrt(w_s_gra)
b_ada = np.sqrt(b_s_gra)
w = w - l_rate * w_grad/w_ada
b = b - l_rate * b_grad/b_ada
可能有编辑错的地方,凑合看吧,实在懒得多写了,这个csdn现在真难用,拼音打一半就自动换行了。