导读
相关pheatmap图,cytoscape网络图,一文打尽。
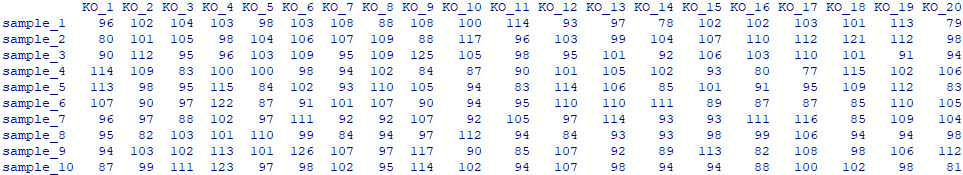
一、模拟输入文件
ko_abun = as.data.frame(matrix(abs(round(rnorm(200, 100, 10))), 10, 20))
colnames(ko_abun) = paste("KO", 1:20, sep="_")
rownames(ko_abun) = paste("sample", 1:10, sep="_")
ko_abun
二、组内相关分析
1 写函数
library(psych)
library(stringr)
correlate = function(other, metabo, route)
{
# 读取方式:check.name=F, row.names=1, header=T
# 计算相关性:
#other = data
#metabo = env
#route="gut"
result=data.frame(print(corr.test(other, metabo, use="pairwise", method="spearman", adjust="fdr", alpha=.05, ci=TRUE, minlength=100), short=FALSE, digits=5))
# FDR矫正
result_raw=data.frame(print(corr.test(other, metabo, use="pairwise", method="spearman", adjust="none", alpha=.05, ci=TRUE, minlength=100), short=FALSE, digits=5))
# 原始P value
# 整理结果
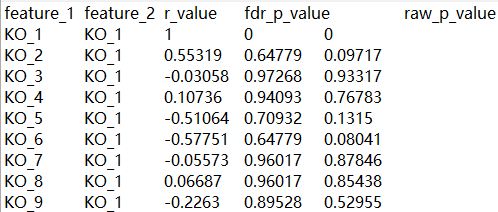
pair=rownames(result) # 行名
result2=data.frame(pair, result[, c(2, 4)]) # 提取信息
# P值排序
# result3=data.frame(result2[order(result2[,"raw.p"], decreasing=F),])
# 格式化结果【将细菌代谢物拆成两列】
result4=data.frame(str_split_fixed(result2$pair, "-", 2), result2[, c(2, 3)], p_value=result_raw[, 4])
colnames(result4)=c("feature_1", "feature_2", "r_value", "fdr_p_value", "raw_p_value")
# 保存提取的结果
write.table(result4, file=paste(route, "Correlation_result.txt", sep="/"), sep="\t", row.names=F, quote=F)
}
2 相关分析
dir.create("Result") # 创建结果目录
correlate(ko_abun, ko_abun, "Result")
三、pheatmap热图
1 写函数
library(reshape2)
library(pheatmap)
correlate_pheatmap = function(infile, route)
{
data=read.table(paste(route, infile, sep="/"), sep="\t", header=T)
data_r=dcast(data, feature_1 ~ feature_2, value.var="r_value")
data_p=dcast(data, feature_1 ~ feature_2, value.var="raw_p_value")
rownames(data_r)=data_r[,1]
data_r=data_r[,-1]
rownames(data_p)=data_p[,1]
data_p=data_p[,-1]
# 剔除不显著的行
del_row = c()
for(i in 1:length(data_p[, 1]))
{
if(all(data_p[i, ] > 0.05))
{
del_row = c(del_row, i)
}
}
# 剔除不显著的列
del_col = c()
for(j in 1:length(data_p[1, ]))
{
if(all(data_p[, j] > 0.05))
{
del_col = c(del_col, j)
}
}
# null值处理
if(is.null(del_row) && !(is.null(del_col)))
{
data_p = data_p[, -del_col]
data_r = data_r[, -del_col]
}else if(is.null(del_col) && !(is.null(del_row)))
{
data_p = data_p[-del_row,]
data_r = data_r[-del_row,]
}else if(is.null(del_row) && is.null(del_col))
{
print("delete none")
}else if(!(is.null(del_row)) && !(is.null(del_col)))
{
data_p = data_p[-del_row, -del_col]
data_r = data_r[-del_row, -del_col]
}
# data_p = data_p[-del_row, -del_col]
# data_r = data_r[-del_row, -del_col]
write.csv(data_p, file=paste(route, "data_p.csv", sep="/"))
write.csv(data_r, file=paste(route, "data_r.csv", sep="/"))
# 用"*"代替<=0.05的p值,用""代替>0.05的相对丰度
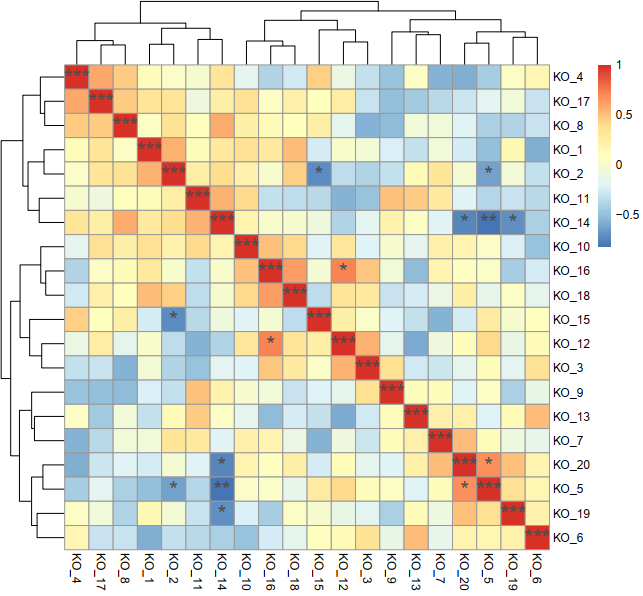
data_mark=data_p
for(i in 1:length(data_p[,1])){
for(j in 1:length(data_p[1,])){
#data_mark[i,j]=ifelse(data_p[i,j] <= 0.05, "*", "")
if(data_p[i,j] <= 0.001)
{
data_mark[i,j]="***"
}
else if(data_p[i,j] <= 0.01 && data_p[i,j] > 0.001)
{
data_mark[i,j]="**"
}
else if(data_p[i,j] <= 0.05 && data_p[i,j] > 0.01)
{
data_mark[i,j]="*"
}
else
{
data_mark[i,j]=""
}
}
}
write.csv(data_mark, file=paste(route, "data_mark.csv", sep="/"))
pheatmap(data_r, display_numbers=data_mark, cellwidth=20, cellheight=20, fontsize_number=18, filename=paste(route, "Correlation_result.pdf", sep="/"))
pheatmap(data_r, display_numbers=data_mark, cellwidth=20, cellheight=20, fontsize_number=18, filename=paste(route, "Correlation_result.png", sep="/"))
}
2 可视化
correlate_pheatmap("Correlation_result.txt", "Result")
3 结果
三个绘图文件和两幅图
四、cytoscape网络图
1 文件准备
先删除自我相关,也就是r值为1的相关。
然后,更新行号。
接着,建feature1-feature2的box。
遍历所有feature2-feature1与box[-1]匹配,如果匹配成功则是重复,记录行号,最后删除。
data = read.table("Result/Correlation_result.txt", sep="\t", header=T)
# 删除自我相关
data = data[data$r_value != 1,]
# 删除重复相关
rownames(data) = 1:nrow(data)
box = paste(data$feature_1, data$feature_2, sep="-")
delete = c()
for(i in 1:nrow(data))
{
tmp = paste(data[i, 2], data[i, 1], sep="-")
box = box[-1]
if(tmp %in% box)
{
delete = c(delete, i)
}
}
data = data[-delete,]
# 画图文件
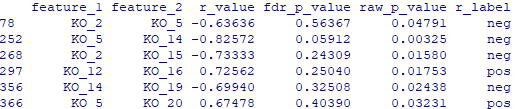
data = data[data$raw_p_value <= 0.05,]
r_label = c()
for(i in 1:nrow(data))
{
if(data[i, 3] < 0)
{
r_label = c(r_label, "neg")
}
else
{
r_label = c(r_label, "pos")
}
}
data$r_label = r_label
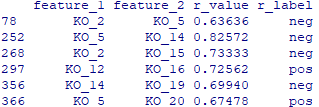
write.table(data, file="Result/input_pre.txt", sep="\t", row.names=F, quote=F)
data$r_value = abs(data$r_value)
input_network = data[, c(1,2,3,6)]
write.table(input_network, file="Result/input_network.txt", sep="\t", row.names=F, quote=F)
2 cytoscape绘图
绘图流程见我的另一篇:Cytoscape绘制相关网络图
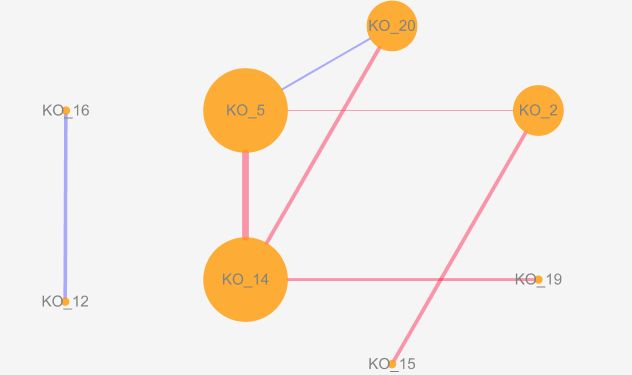
网络图结果如下:
圆圈表示KO,相关越多degree,圆越大
先粗细表示|r|,红色负相关,蓝色正相关