今天继续 跟着Nature Communications学画图系列第二篇。学习R语言基础绘图函数画散点图。
对应的 Nature Communications 的论文是 Fecal pollution can explain antibiotic resistance gene abundances in anthropogenically impacted environments
这篇论文数据分析和可视化的部分用到的数据和代码全部放到了github上 https://github.com/karkman/crassphage_project
非常好的R语言学习素材。
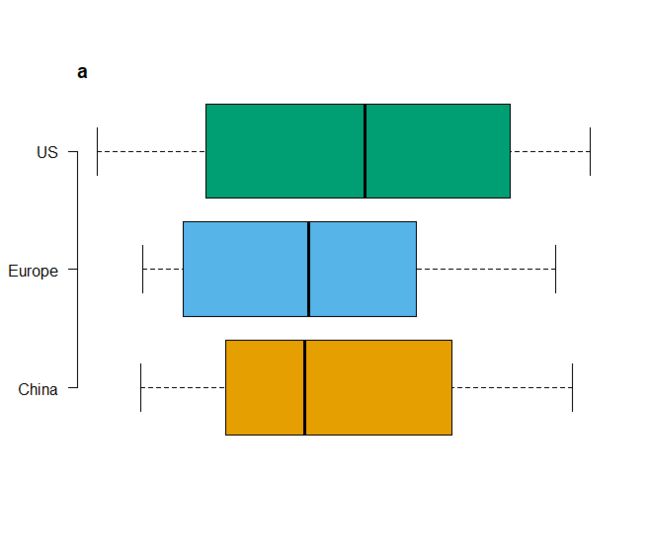
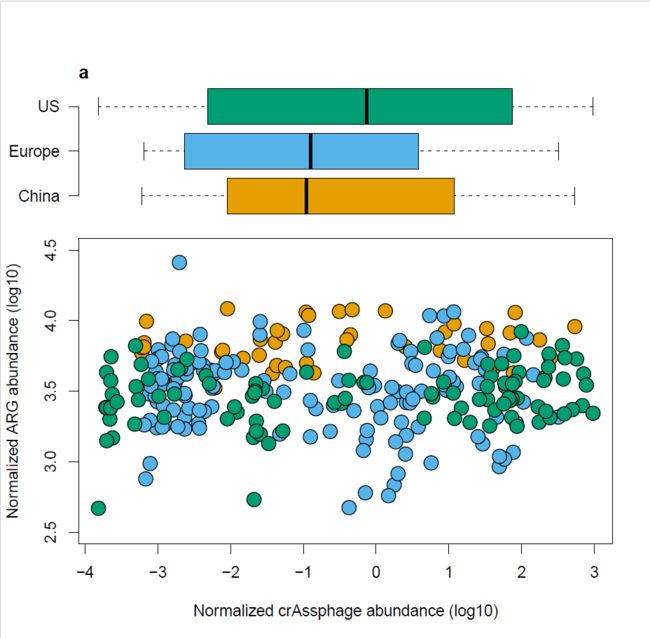
今天学习Figure1中被红色框线圈住的散点图
第一部分先收一下上一篇文章的尾
https://mp.weixin.qq.com/s/dFM1bJoEGPjWhEFTaQ0Krg
跟着Nature Communications学画图~ Figure1 ~基础绘图函数箱线图
这篇文章中有人留言说 和原图不是很像,因为配色没有按照论文中提供的代码来。
下面是完全重复论文中的代码
cols <- c("#E69F00", "#56B4E9", "#009E73")
boxplot(log10(rel_crAss)~country,data=HMP,col=cols,
axes=F,xlab=NULL,ylab=NULL,
horizontal = T)
axis(2,at=c(1,2,3),labels=c("China", "Europe", "US"),las=1)
title("a",adj=0,line=0)
第二部分 基础绘图函数散点图
- 读入数据
HMP<-read.table("data/HMP.txt")
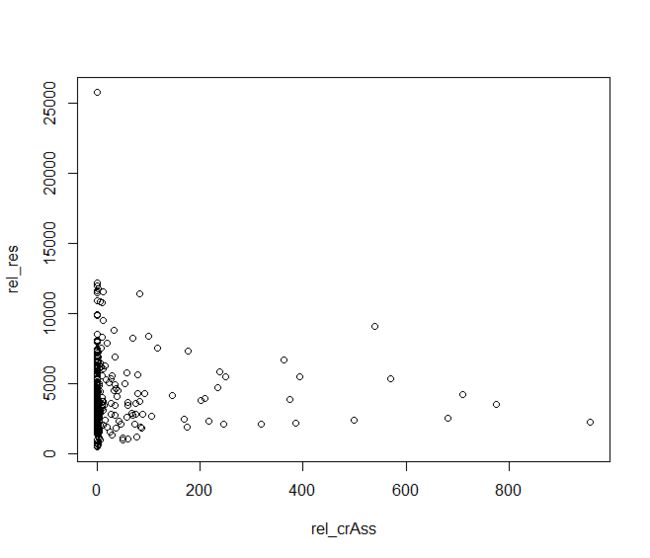
- 最基本的散点图
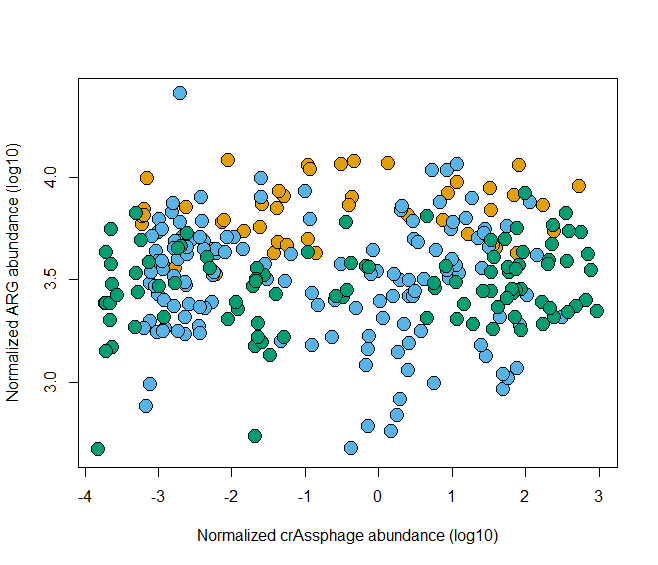
plot(rel_res~rel_crAss,data=HMP)
画图用plot()函数,需要指定画图用到的变量y和x,还有画图用到的数据data
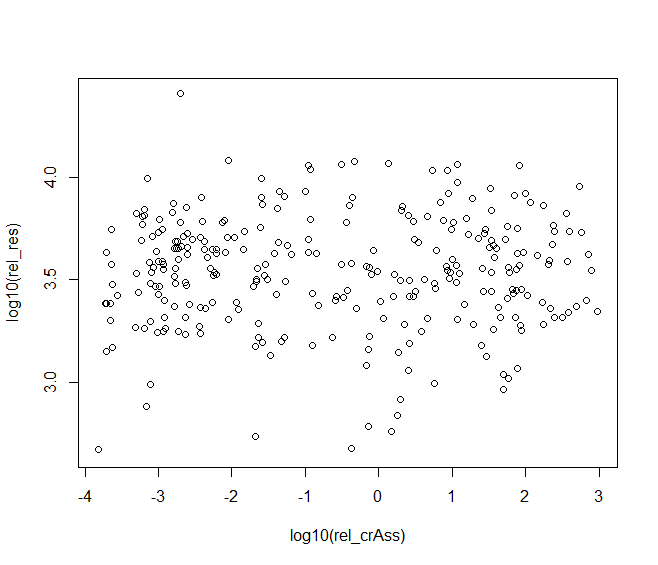
原始代码分别对 rel_res 和 rel_crAss取了log10
plot(log10(rel_res)~log10(rel_crAss),data=HMP)
取log10以后可以看到散点分布的更加均匀了。
接下来就是对图进行美化了
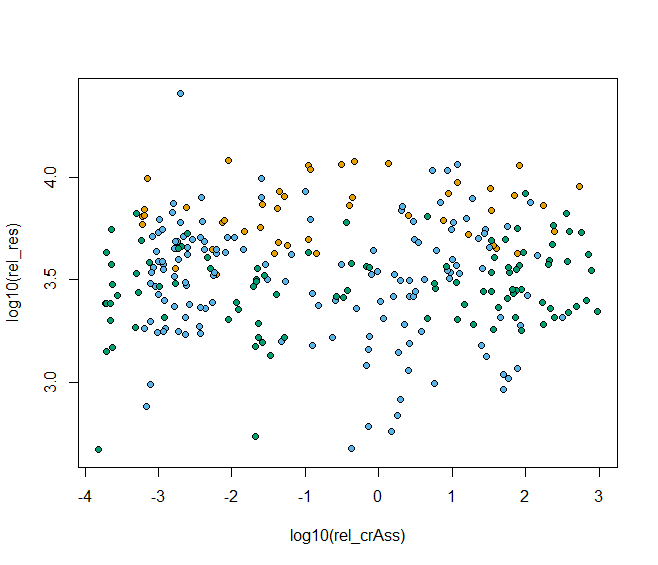
- 按照国家分组填充颜色
cols <- c("#E69F00", "#56B4E9", "#009E73", "#F0E442", "#0072B2", "#D55E00", "#CC79A7")
plot(log10(rel_res)~log10(rel_crAss), data=HMP,
bg=cols[as.factor(HMP$country)],pch=21)
- 更改点的大小
plot(log10(rel_res)~log10(rel_crAss), data=HMP,
bg=cols[as.factor(HMP$country)],pch=21,cex=2)
- 更改x轴和y轴的标签
plot(log10(rel_res)~log10(rel_crAss), data=HMP, bg=cols[as.factor(HMP$country)], pch=21,
ylab = "Normalized ARG abundance (log10)",
xlab="Normalized crAssphage abundance (log10)", cex=2)
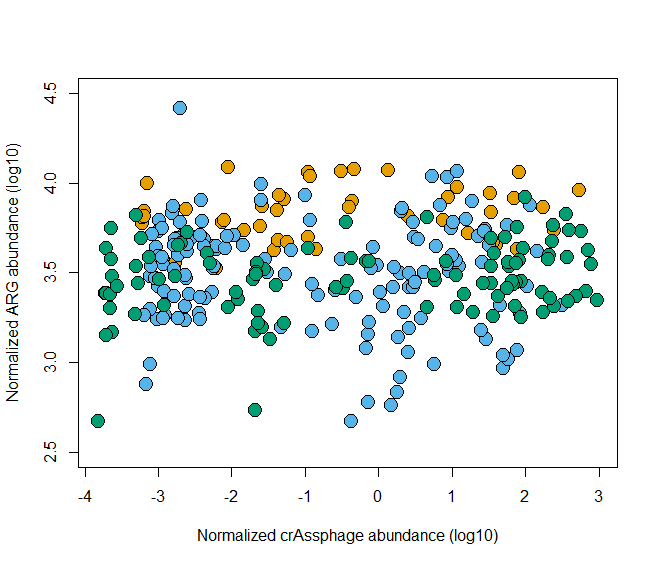
- 更改坐标轴的范围
plot(log10(rel_res)~log10(rel_crAss), data=HMP,
bg=cols[as.factor(HMP$country)], pch=21,
ylab = "Normalized ARG abundance (log10)",
xlab="Normalized crAssphage abundance (log10)",
cex=2,
ylim=c(2.5, 4.5))
接下来将箱线图和散点图按照上下拼接到一起,用到的是 具体可以参考链接 欢迎大家关注我的公众号par(fig=c(a,b,c,d)),这里需要满足 a
https://blog.csdn.net/qingchongxinshuru/article/details/52004182par(fig=c(0,1,0,0.75))
plot(log10(rel_res)~log10(rel_crAss), data=HMP,
bg=cols[as.factor(HMP$country)], pch=21,
ylab = "Normalized ARG abundance (log10)",
xlab="Normalized crAssphage abundance (log10)",
cex=2,
ylim=c(2.5, 4.5))
par(fig=c(0,1,0.5,1),new=T)
boxplot(log10(rel_crAss)~country,data=HMP,col=cols,
axes=F,xlab=NULL,ylab=NULL,
horizontal = T)
axis(2,at=c(1,2,3),labels=c("China", "Europe", "US"),las=1)
title("a",adj=0,line=0)
小明的数据分析笔记本