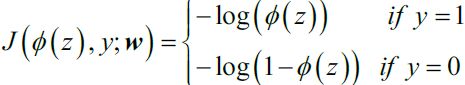cp15_Classifying Images with Deep Convolutional NN_Loss_Cross Entropy_ax.text_mnist_ CelebA_Colab_ck
In the previous chapter, we looked in depth at different aspects of the TensorFlow API, became familiar with tensors, naming variables, and operators, and learned how to work with variable scopes. In this chapter, we'll now learn about Convolutional Neural Networks (CNNs), and how we can implement CNNs in TensorFlow. We'll also take an interesting journey in this chapter as we apply this type of deep neural network architecture to image classification.
So we'll start by discussing the basic building blocks of CNNs, using a bottom-up approach. Then we'll take a deeper dive into the CNN architecture and how to implement deep CNNs in TensorFlow. Along the way we'll be covering the following topics:
- Understanding convolution operations in one and two dimensions
- Learning about the building blocks of CNN architectures
- Implementing deep convolutional neural networks in TensorFlow
Building blocks of convolutional neural networks
Convolutional neural networks, or CNNs, are a family of models that were inspired by how the visual cortex of human brain works when recognizing objects.
The development of CNNs goes back to the 1990's, when Yann LeCun and his colleagues proposed a novel neural network architecture for classifying handwritten digits from images (Handwritten Digit Recognition with a Back-Propagation Network, Y LeCun, and others, 1989, published at Neural Information Processing Systems.(NIPS) conference).
Due to the outstanding performance of CNNs for image classification tasks, they have gained a lot of attention and this led to tremendous improvements in machine learning and computer vision applications.
In the following sections, we next see how CNNs are used as feature extraction engines, and then we'll delve into the theoretical definition of convolution and computing convolution in one and two dimensions.
Understanding CNNs and learning feature hierarchies
Successfully extracting salient[ˈseɪliənt]显著的, 重要的, 突出的, 主要的 (relevant) features is key to the performance of any machine learning algorithm, of course, and traditional machine learning models rely on input features that may come from a domain expert, or are based on computational feature extraction techniques. Neural networks are able to automatically learn the features from raw data that are most useful for a particular task. For this reason, it's common to consider a neural network as a feature extraction engine: the early layers (those right after the input layer) extract low-level features.
Multilayer neural networks, and in particular, deep convolutional neural networks, construct a so-called feature hierarchy by combining the low-level features in a layer-wise fashion to form high-level features. For example, if we're dealing with
images, then low-level features, such as edges and blobs边缘和斑点, are extracted from the earlier layers, which are combined together to form high-level features – as object shapes like a building, a car, or a dog.
As you can see in the following image, a CNN computes feature maps from an input image, where each element comes from a local patch of pixels in the input image: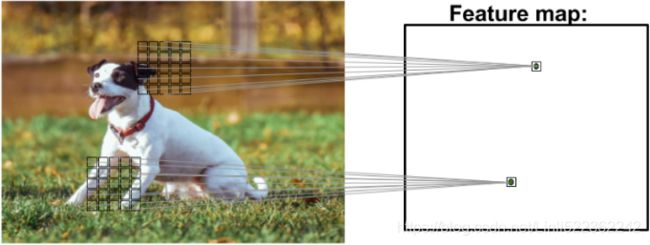
This local patch of pixels is referred to as the local receptive field. CNNs will usually perform very well for image-related tasks, and that's largely due to two important ideas:
- Sparse-connectivity: A single element in the feature map is connected to only a small patch of pixels. (This is very different from connecting to the whole input image, in the case of perceptrons. You may find it useful to look back and compare how we implemented a fully connected network that connected to the whole image, in Chapter 12, Implementing a Multilayer Artificial Neural Network from Scratch.)
- Parameter-sharing: The same weights are used for different patches of the input image. As a direct consequence of these two ideas, the number of weights (parameters) in the network decreases dramatically, and we see an improvement in the ability to capture salient features. Intuitively, it makes sense that nearby pixels are probably more relevant to each other than pixels that are far away from each other.
Typically, CNNs are composed of several Convolutional (conv) layers and subsampling (also known as Pooling (P)) layers that are followed by one or more Fully Connected (FC) layers at the end. The fully connected layers are essentially a multilayer perceptron, where every input unit i is connected to every output unit j with weight ![]() (which we learned about in Chapter 12, Implementing a Multilayer Artificial Neural Network from Scratch).
(which we learned about in Chapter 12, Implementing a Multilayer Artificial Neural Network from Scratch).
Please note that subsampling layers, commonly known as pooling layers, do not have any learnable parameters; for instance, there are no weights or bias units in pooling layers. However, both convolution and fully connected layers have such weights and biases.
In the following sections, we'll study convolutional and pooling layers in more detail and see how they work. To understand how convolution operations work, let's start with a convolution in one dimension before working through the typical two dimensional cases as applications for two-dimensional images later.
Performing discrete convolutions
A discrete convolution (or simply convolution) is a fundamental operation in a CNN. Therefore, it's important to understand how this operation works. In this section, we'll learn the mathematical definition and discuss some of the naive
algorithms to compute convolutions of two one-dimensional vectors or two twodimensional matrices.
Please note that this description is solely for understanding how a convolution works. Indeed, much more efficient implementations of convolutional operations already exist in packages such as TensorFlow, as we will see later in this chapter.
Note
Mathematical notation
In this chapter, we will use subscripts to denote the size of a multidimensional array; for example, ![]() is a two-dimensional array of size
is a two-dimensional array of size ![]() . We use brackets
. We use brackets ![]() to denote the indexing of a multidimensional array. For example,
to denote the indexing of a multidimensional array. For example, ![]() means the element at index
means the element at index ![]() of matrix
of matrix ![]() . Furthermore, note that we use a special symbol * to denote the convolution operation between two vectors or matrices, which is not to be confused with the multiplication operator * in Python.
. Furthermore, note that we use a special symbol * to denote the convolution operation between two vectors or matrices, which is not to be confused with the multiplication operator * in Python.
#############################################################
9.1 The Convolution Operation
In its most general form, convolution is an operation on two functions of a real-valued argument. To motivate the definition of convolution, we start with examples of two functions we might use.
Suppose we are tracking the location of a spaceship with a laser sensor. Our laser sensor provides a single output x(t), the position of the spaceship at time t. Both x and t are real-valued, i.e., we can get a different reading from the laser sensor at any instant in time.
Now suppose that our laser sensor is somewhat noisy. To obtain a less noisy estimate of the spaceship’s position, we would like to average together several measurements测量结果. Of course, more recent measurements are more relevant, so we will
want this to be a weighted average that gives more weight to recent measurements. We can do this with a weighting function w(a), where a is the age of a measurement![]() . If we apply such a weighted average operation at every moment, we obtain a new function providing s a smoothed estimate of the position of the spaceship:
. If we apply such a weighted average operation at every moment, we obtain a new function providing s a smoothed estimate of the position of the spaceship: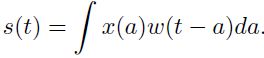 (9.1)
(9.1)
This operation is called convolution. The convolution operation is typically denoted with an asterisk:![]() (9.2)
(9.2)
In our example, w needs to be a valid probability density function, or the output is not a weighted average. Also, w needs to be 0 for all negative arguments, or it will look into the future, which is presumably beyond our capabilities. These
limitations are particular to our example though. In general, convolution is defined for any functions for which the above integral is defined, and may be used for other purposes besides taking weighted averages.
In convolutional network terminology, the first argument (in this example, the function x) to the convolution is often referred to as the input and the second argument (in this example, the function w) as the kernel. The output is sometimes referred to as the feature map.
In our example, the idea of a laser sensor that can provide measurements at every instant in time is not realistic. Usually, when we work with data on a computer, time will be discretized, and our sensor will provide data at regular intervals. In our example, it might be more realistic to assume that our laser provides a measurement测量结果 once per second. The time index t can then take on only integer values. If we now assume that x and w are defined only on integer t, we can define the discrete convolution: (9.3)
(9.3)
In machine learning applications, the input is usually a multidimensional array of data and the kernel is usually a multidimensional array of parameters that are adapted by the learning algorithm. We will refer to these multidimensional arrays as tensors. Because each element of the input and kernel must be explicitly stored separately, we usually assume that these functions are zero everywhere but(except) the finite set of points for which we store the values. This means that in practice we can implement the infinite summation as a summation over a finite number of array elements.
#############################################################
Performing a discrete convolution in one dimension
Let's start with some basic definitions and notations we are going to use. A discrete convolution for two one-dimensional vectors x and w is denoted by ![]() , in which vector x is our input (sometimes called signal) and w is called the filter or kernel. A discrete convolution is mathematically defined as follows:
, in which vector x is our input (sometimes called signal) and w is called the filter or kernel. A discrete convolution is mathematically defined as follows:
Here, the brackets [ ] are used to denote the indexing for vector elements. The index i runs through each element of the output vector y. There are two odd things in the preceding formula that we need to clarify: ![]() to
to ![]() indices and negative indexing for x.
indices and negative indexing for x.
Tip
Cross-correlation互相关
Cross-correlation (or simply correlation) between an input vector and a filter is denoted by ![]() and is very much like a sibling for a discrete convolution with a small difference; the difference is that in cross-correlation, the multiplication is performed in the same direction. Therefore, it is not required to rotate the filter matrix w in each dimension. Mathematically, cross-correlation is defined as follows:
and is very much like a sibling for a discrete convolution with a small difference; the difference is that in cross-correlation, the multiplication is performed in the same direction. Therefore, it is not required to rotate the filter matrix w in each dimension. Mathematically, cross-correlation is defined as follows:
The same rules for padding and stride may be applied to cross-correlation as well. The first issue where the sum runs through indices from ![]() to
to ![]() seems odd mainly because in machine learning applications, we always deal with finite feature vectors. For example, if x has 10 features with indices
seems odd mainly because in machine learning applications, we always deal with finite feature vectors. For example, if x has 10 features with indices ![]() , then indices
, then indices ![]() and
and ![]() are out of bounds for x. Therefore, to correctly compute the summation shown in the preceding formula, it is assumed that x and w are filled with zeros. This will result in an output vector y that also has infinite size with lots of zeros as well. Since this is not useful in practical situations, x is padded only with a finite number of zeros.
are out of bounds for x. Therefore, to correctly compute the summation shown in the preceding formula, it is assumed that x and w are filled with zeros. This will result in an output vector y that also has infinite size with lots of zeros as well. Since this is not useful in practical situations, x is padded only with a finite number of zeros.
This process is called zero-padding or simply padding. Here, the number of zeros padded on each side is denoted by p. An example padding of a one-dimensional vector x is shown in the following figure: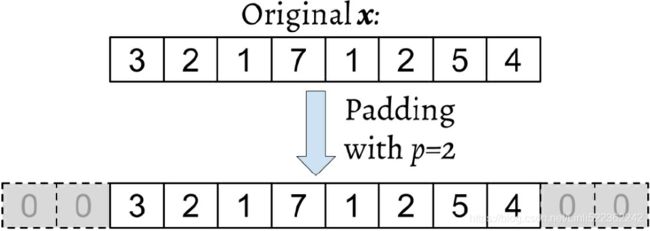
Let's assume that the original input x and filter w have n and m elements, respectively, where ![]() . Therefore, the padded vector
. Therefore, the padded vector ![]() has size n + 2p. Then, the practical formula for computing a discrete convolution will change to the following:
has size n + 2p. Then, the practical formula for computing a discrete convolution will change to the following:![]() (0 <= k <= m-1) <==
(0 <= k <= m-1) <== 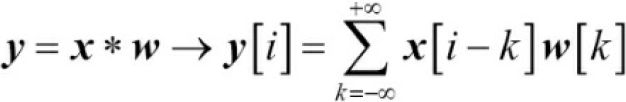
Now that we have solved the infinite index issue, the second issue is indexing x with i + m - k. The important point to notice here is that x and w are indexed in different directions in this summation. For this reason, we can flip one of those vectors, x or w, after they are padded. Then, we can simply compute their dot product.
Let's assume we flip the filter w to get the rotated filter ![]() . Then, the dot product
. Then, the dot product 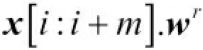 is computed to get one element
is computed to get one element ![]() , where
, where ![]() is a patch of x with size m. (hidden stride s=1; if s>1, then x[i*s:i*s+m])
is a patch of x with size m. (hidden stride s=1; if s>1, then x[i*s:i*s+m])
This operation is repeated like in a sliding滑动 window approach to get all the output elements. The following figure provides an example with x = (3,2,1,7,1,2,5,4) and 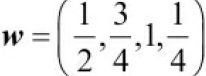 so that the first three output elements are computed:
so that the first three output elements are computed: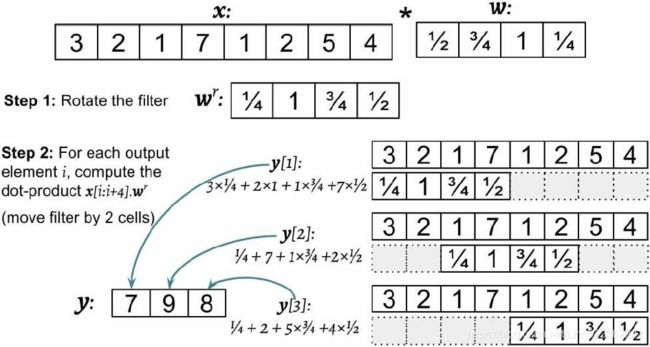
You can see in the preceding example that the padding size is zero (p = 0). Notice that the rotated filter ![]() is shifted by two cells each time we shift. This shift is another hyperparameter of a convolution, the stride s. In this example, the stride is two, s = 2 (<2). Note that the stride has to be a positive number smaller than the size of the input vector. We'll talk more about padding and strides in the next section!
is shifted by two cells each time we shift. This shift is another hyperparameter of a convolution, the stride s. In this example, the stride is two, s = 2 (<2). Note that the stride has to be a positive number smaller than the size of the input vector. We'll talk more about padding and strides in the next section!
The effect of zero-padding in a convolution
So far here, we've used zero-padding in convolutions to compute finite-sized output vectors. Technically, padding can be applied with any ![]() . Depending on the choice p, boundary cells may be treated differently than the cells located in the middle of x.
. Depending on the choice p, boundary cells may be treated differently than the cells located in the middle of x.
Now consider an example where n = 5, m = 3 (input x and filter w have n and m elements).
Then, p = 0, x[0] is only used in computing one output element (for instance, y[0], and stride s =1),
while x[1] is used in the computation of two output elements (for instance, y[0] and y[1], stride s =1). So, you can see that this different treatment of elements of x can artificially put more emphasis on the middle element, x[1], since it has appeared in most computations.
We can avoid this issue if we choose p = 2, in which case, each element of x will be involved in computing three elements of y.
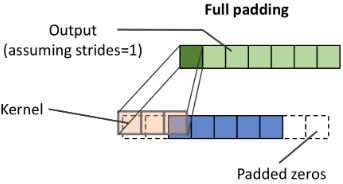
Furthermore, the size of the output y also depends on the choice of the padding strategy we use. There are three modes of padding that are commonly used in practice: full, same, and valid:
- In the full mode, the padding parameter p is set to p = m - 1. Full padding increases the dimensions of the output; thus, it is rarely used in convolutional neural network architectures.
- Same padding is usually used if you want to have the size of the output the same as the input vector x. In this case, the padding parameter p is computed according to the filter size, along with the requirement that the input size and output size are the same.
- Finally, computing a convolution in the valid mode refers to the case where p = 0 (no padding).
The following figure illustrates the three different padding modes for a simple 5 x 5 pixel input with a kernel size of 3 x 3 and a stride of 1:
###############################################################

###############################################################
The most commonly used padding mode in convolutional neural networks is same padding. One of its advantages over the other padding modes is that same padding preserves the height and width of the input images or tensors, which makes designing a network architecture more convenient.
One big disadvantage of the valid padding versus full and same padding, for example, is that the volume of the tensors would decrease substantially in neural networks with many layers, which can be detrimental [ˌdetrɪˈmentl]有害的 to the network performance.
In practice, it is recommended that you preserve the spatial size空间大小 using same padding for the convolutional layers and decrease the spatial size via pooling layers instead. As for the full padding, its size results in an output larger than the input size. Full padding is usually used in signal processing applications where it is important to minimize boundary effects. However, in deep learning context, boundary effect is not usually an issue, so we rarely see full padding.
Determining the size of the convolution output
The output size of a convolution is determined by the total number of times that we shift the filter w along the input vector x. Let's assume that the input vector has size n and the filter is of size m. Then, the size of the output resulting from ![]() with padding p and stride s is determined as follows:
with padding p and stride s is determined as follows: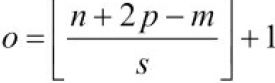
Here, ![]() denotes the floor operation
denotes the floor operation
Tip
The floor operation returns the largest integer that is equal or smaller to the input, for example:![]()
Consider the following two cases:
- Compute the output size for an input vector of size 10 with a convolution kernel of size 5, padding 2, and stride 1:
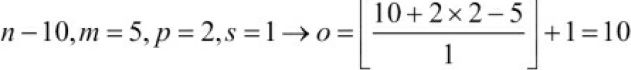
(Note that in this case, the output size turns out to be the same as the input; therefore, we conclude this as mode='same')
- How can the output size change for the same input vector(input vector of size 10), but have a kernel of size 3, and stride 2?
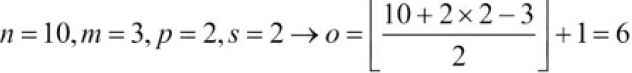
If you are interested to learn more about the size of the convolution output, we recommend the manuscript A guide to convolution arithmetic for deep learning, Vincent Dumoulin and Francesco Visin, 2016, which is freely available at
https://arxiv.org/abs/1603.07285.
Finally, in order to learn how to compute convolutions in one dimension, a naïve implementation is shown in the following code block, and the results are compared with the numpy.convolve function. The code is as follows:
Performing a discrete convolution in 1D
import tensorflow as tf
import numpy as np
def conv1d(x,w, p=0, s=1):
w_rot = np.array(w[::-1])
x_padded = np.array(x)
if p>0 :
zero_pad = np.zeros(shape=p)
x_padded = np.concatenate([zero_pad, x_padded, zero_pad]) #len(x_padded) == n+2p
res = []
# floor( (n+2p - m) /s) + 1
for i in range(0, int( ( len(x_padded) -len(w_rot) )/s )+ 1):######## corrected
#x_padded[i:i+m] * w_rot #hidden: s=1, but here s>=1
res.append( np.sum(x_padded[ i*s:i*s+w_rot.shape[0] ] * w_rot) )# corrected
return np.array(res)
## Testing:
x = [3,2,1,7,1,2,5,4] # [1,3,2,4,5,6, 1,3]
w = [1/2,3/4,1,1/4] # [1,0,3,1,2]
print( 'Conv1d Implementation: ', conv1d(x,w, p=0, s=2) )
print( 'Numpy Results: ', np.convolve(x,w, mode='valid')) #s=1Conv1d Implementation: [7. 9. 8.]
Numpy Results: [7. 7.25 9. 6.75 8. ] #since the stride of np.convolve is alway 1, but here the stride=2
## Testing:
x = [1,3,2,4,5,6, 1,3]
w = [1,0,3,1,2]
print( 'Conv1d Implementation: ', conv1d(x,w, p=2, s=1) )
print( 'Numpy Results: ', np.convolve(x,w, mode='same')) #s=1![]()
So far, here, we have explored the convolution in 1D. We started with 1D case to make the concepts easier to understand. In the next section, we will extend this to two dimensions.
Performing a discrete convolution in 2D
The concepts you learned in the previous sections are easily extendible to two dimensions. When we deal with two-dimensional input, such as a matrix ![]() and the filter matrix
and the filter matrix 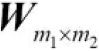 , where
, where ![]() and
and ![]() , then the matrix
, then the matrix ![]() is the result of 2D convolution of X with W. This is mathematically defined as follows:
is the result of 2D convolution of X with W. This is mathematically defined as follows: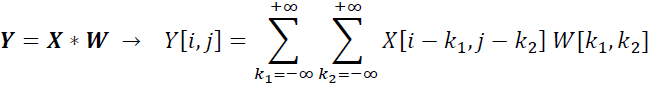
Notice that if you omit one of the dimensions, the remaining formula is exactly the same as the one we used previously to compute the convolution in 1D. In fact, all the previously mentioned techniques, such as zero-padding, rotating the filter matrix,
and the use of strides, are also applicable to 2D convolutions, provided that they are extended to both the dimensions independently. The following example illustrates the computation of a 2D convolution between an input matrix ![]() , a kernel matrix
, a kernel matrix ![]() , padding
, padding ![]() , and stride
, and stride ![]() . According to the specified padding, one layer of zeros are padded on each side of the input matrix, which results in the padded matrix
. According to the specified padding, one layer of zeros are padded on each side of the input matrix, which results in the padded matrix ![]() , as follows:
, as follows:
With the preceding filter, the rotated filter will be:
Note that this rotation is not the same as the transpose matrix. To get the rotated filter in NumPy, we can write W_rot=W[::-1,::-1]. Next, we can shift the rotated filter matrix along the padded input matrix ![]() like a sliding window and
like a sliding window and
compute the sum of the element-wise product, which is denoted by the ![]() operator in the following figure:
operator in the following figure: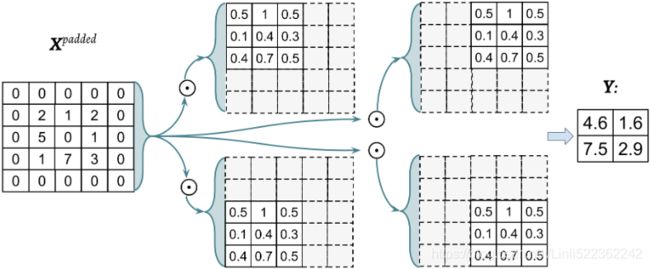
The result will be the 2 x 2 matrix Y.
###################################################
In each channel, the Input * Kernel is using element-wise multiplication, then sum all elements in the result of multiplication
next step, use the intermediate outputs do element-wise addition
Figure 5: A 2D Convolution with a 3x3 kernel applied to an 3 channel RGB input of size 5x5 to give output of 3x3.
###################################################
Let's also implement the 2D convolution according to the naïve algorithm described. The scipy.signal package provides a way to compute 2D convolution via the scipy.signal.convolve2d function:
import scipy.signal
def conv2d(X,W, p=(0,0), s=(1,1)):
W_rot = np.array(W)[::-1, ::-1]
X_orig = np.array(X)
n1 = X_orig.shape[0] + 2*p[0]
n2 = X_orig.shape[1] + 2*p[1]
X_padded = np.zeros( shape=(n1,n2) )
X_padded[ p[0]:p[0]+X_orig.shape[0], # select rows
p[1]:p[1]+X_orig.shape[1] # select columns
] = X_orig
res = [] # floor( ( n+2p - m) /s) +1
for i in range(0, int( ( X_padded.shape[0]-W_rot.shape[0])/s[0])+1 ):
res.append([])
for j in range(0, int( ( X_padded.shape[1]-W_rot.shape[1])/s[1])+1 ):
X_sub = X_padded[ i*s[0]:i*s[0]+W_rot.shape[0],
j*s[1]:j*s[1]+W_rot.shape[1]
]
# X_sub*W_rot : element-wise multiplication, then sum all elements in the result
# for example:
# X_sub:
# [[0. 0. 0.]
# [0. 2. 1.]
# [0. 5. 0.]]
# W_rot:
# [[0.5 1. 0.5]
# [0.1 0.4 0.3]
# [0.4 0.7 0.5]]
# X_sub*W_rot:
# [[0. 0. 0. ]
# [0. 0.8 0.3]
# [0. 3.5 0. ]]
# np.sum(X_sub*W_rot): 4.6
res[-1].append( np.sum(X_sub*W_rot) )#append the result of summation on last row
return( np.array(res) )
X = [[2,1,2],
[5,0,1],
[1,7,3]
]
W = [[0.5,0.7,0.4],
[0.3,0.4,0.1],
[0.5, 1, 0.5]
]
print('Conv2d Implementation:\n',
conv2d( X,W, p=(1,1), s=(2,2) )
)
print('Scipy Results:\n', scipy.signal.convolve2d(X,W,mode='same'))Conv2d Implementation:
[ [4.6 1.6]
[7.5 2.9]
]
Scipy Results:
[ [ 4.6 3.7 1.6]
[ 8.7 10.6 7.8]
[ 7.5 6.8 2.9]
] ##since the stride of scipy.signal.convolve2d is alway 1, but here the stride=2
X = [[1, 3, 2, 4],
[5, 6, 1, 3],
[1, 2, 0, 2],
[3, 4, 3, 2]
]
W = [[1, 0, 3],
[1, 2, 1],
[0, 1, 1]
]
print('Conv2d Implementation:\n',
conv2d( X,W, p=(1,1), s=(1,1) )
)
print('Scipy Results:\n', scipy.signal.convolve2d(X,W,mode='same'))Tip
We provided a naïve implementation to compute a 2D convolution for the purpose of understanding the concepts. However, this implementation is very inefficient in terms of memory requirements and computational complexity. Therefore, it should not be used in real-world neural network applications.
In recent years, much more efficient algorithms have been developed that use the Fourier transformation for computing convolutions. It is also important to note that in the context of neural networks, the size of a convolution kernel is usually much smaller than the size of the input image. For example, modern CNNs usually use kernel sizes such as 1 x 1, 3 x 3, or 5 x 5, for which efficient algorithms have been designed that can carry out the convolutional operations much more efficiently, such as the Winograd's Minimal Filtering algorithm. These algorithms are beyond the scope of this book, but if you are interested to learn more, you can read the manuscript: Fast Algorithms for Convolutional Neural Networks, Andrew Lavin and
Scott Gray, 2015, which is freely available at (https://arxiv.org/abs/1509.09308).
In the next section, we will discuss subsampling, which is another important operation often used in CNNs.
Subsampling layers
Subsampling is typically applied in two forms of pooling operations in convolutional neural networks: max-pooling and mean-pooling (also known as average-pooling).
The pooling layer is usually denoted by ![]() . Here, the subscript determines the size of the neighborhood (the number of adjacent pixels in each dimension), where the max or mean operation is performed. We refer to such a neighborhood as the pooling size.
. Here, the subscript determines the size of the neighborhood (the number of adjacent pixels in each dimension), where the max or mean operation is performed. We refer to such a neighborhood as the pooling size.
The operation is described in the following figure. Here, max-pooling takes the maximum value from a neighborhood of pixels, and mean-pooling computes their average: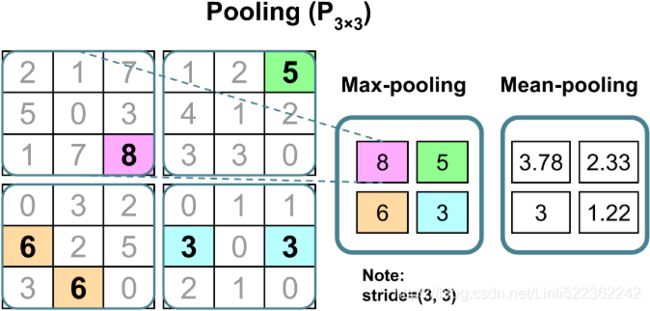
The advantage of pooling is twofold:
- Pooling (max-pooling) introduces some sort of local invariance. This means that small changes in a local neighborhood do not change the result of maxpooling. Therefore, it helps generate features that are more robust to noise in the input data. See the following example that shows max-pooling of two different input matrices
 and
and  results in the same output:
results in the same output: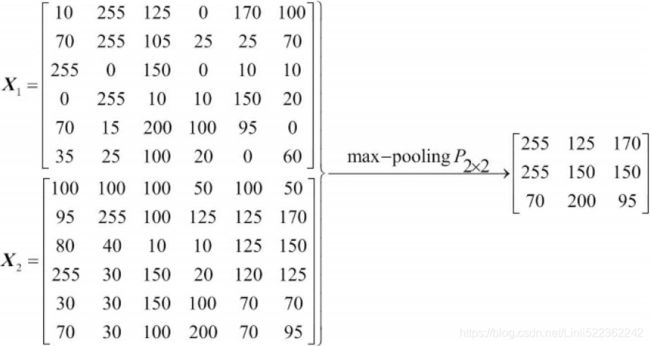
- Pooling decreases the size of features, which results in higher computational efficiency. Furthermore, reducing the number of features may reduce the degree of overfitting as well.
Note
Traditionally, pooling is assumed to be nonoverlapping. Pooling is typically performed on nonoverlapping neighborhoods, which can be done by setting the stride parameter equal to the pooling size ![]() . For example, a nonoverlapping
. For example, a nonoverlapping
pooling layer requires a stride parameter ![]() .
.
On the other hand, overlapping pooling occurs if the stride is smaller than pooling size. An example where overlapping pooling is used in a convolutional network is described in ImageNet Classification with Deep Convolutional Neural Networks, A. Krizhevsky, I. Sutskever, and G. Hinton, 2012, which is freely available as a manuscript at https://papers.nips.cc/paper/4824-imagenetclassification-with-deep-convolutional-neural-networks.
Putting everything together to build a CNN
So far, we've learned about the basic building blocks of convolutional neural networks. The concepts illustrated in this chapter are not really more difficult than traditional multilayer neural networks. Intuitively, we can say that the most important operation in a traditional neural network is the matrix-vector multiplication.
For instance, we use matrix-vector multiplications to pre-activations (or net input) as in ![]() . Here, x is a column vector representing pixels, and W is the weight matrix connecting the pixel inputs to each hidden unit. In a convolutional neural network, this operation is replaced by a convolution operation, as in
. Here, x is a column vector representing pixels, and W is the weight matrix connecting the pixel inputs to each hidden unit. In a convolutional neural network, this operation is replaced by a convolution operation, as in ![]() , where X is a matrix representing the pixels in a [height x width] arrangement. In both cases, the pre-activations are passed to an activation function to obtain the activation of a hidden unit
, where X is a matrix representing the pixels in a [height x width] arrangement. In both cases, the pre-activations are passed to an activation function to obtain the activation of a hidden unit ![]() , where
, where ![]() is the activation function. Furthermore, recall that subsampling( OR pooling) is another building block of a convolutional neural network, which may appear in the form of pooling, as we described in the previous section.
is the activation function. Furthermore, recall that subsampling( OR pooling) is another building block of a convolutional neural network, which may appear in the form of pooling, as we described in the previous section.
Working with multiple input or color channels
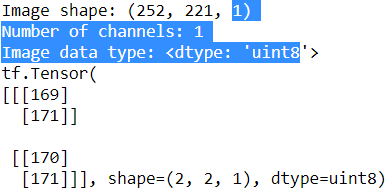
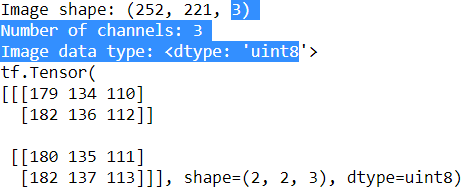
An input sample to a convolutional layer may contain one or more 2D arrays or matrices with dimensions ![]() (for example, the image height and width in pixels). These
(for example, the image height and width in pixels). These ![]() matrices are called channels. Therefore, using multiple channels as input to a convolutional layer requires us to use a rank-3 tensor or a three-dimensional array:
matrices are called channels. Therefore, using multiple channels as input to a convolutional layer requires us to use a rank-3 tensor or a three-dimensional array: ![]() , where
, where ![]() is the number of input channels. For example, let's consider images as input to the first layer of a CNN. If the image is colored and uses the RGB color mode, then
is the number of input channels. For example, let's consider images as input to the first layer of a CNN. If the image is colored and uses the RGB color mode, then ![]() (for the red, green, and blue color channels in RGB). However, if the image is in grayscale, then we have
(for the red, green, and blue color channels in RGB). However, if the image is in grayscale, then we have ![]() because there is only one channel with the grayscale pixel intensity values.
because there is only one channel with the grayscale pixel intensity values.
Tip
When we work with images, we can read images into NumPy arrays using the 'uint8' (unsinged 8-bit integer) data type to reduce memory usage compared to 16-bit, 32-bit, or 64-bit integer types, for example. Unsigned 8-bit integers take values
in the range [0, 255], which are sufficient to store the pixel information in RGB images, which also take values in the same range.
Next, let's look at an example of how we can read in an image into our Python session using SciPy. However, please note that reading images with SciPy requires that you have the Python Imaging Library (PIL) package installed. We can install
Pillow (https://python-pillow.org), a more user-friendly fork of PIL, to satisfy those requirements, as follows:
pip install pillow
Once Pillow is installed, we can use the imread function from the scipy.misc module to read an RGB image (this example image is located in the code bundle folder that is provided with this chapter at https://github.com/rasbt/python-machinelearning-
book-2nd-edition/tree/master/code/ch15):
# import scipy.misc
# img = scipy.misc.imread('example-image.png', mode='RGB')
# AttributeError: module 'scipy.misc' has no attribute 'imread'
# let's use tf.io.read_file
img_raw = tf.io.read_file('example-image-gray.png')#img_raw = tf.io.read_file('example-image.png')#INFO-BOX: The rank of a grayscale image for input to a CNN
img_raw = tf.io.read_file('example-image-gray.png')
# ==> Now that we have familiarized ourselves with the structure of input data, the next question is how can we incorporate multiple input channels in the convolution operation that we discussed in the previous sections?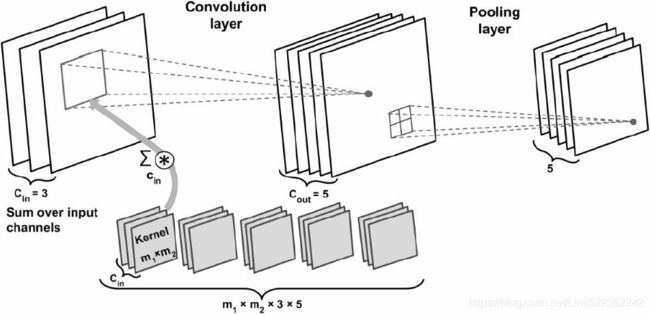
The answer is very simple: we perform the convolution operation for each channel separately and then add the results together using the matrix summation. The convolution associated with each channel (c) has its own kernel matrix as ![]() . The total pre-activation result is computed in the following formula:
. The total pre-activation result is computed in the following formula: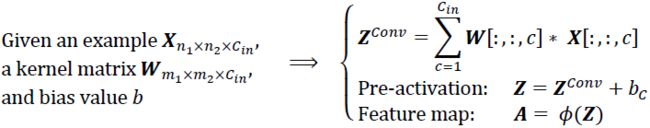
The final result, A, is called a feature map.
Usually, a convolutional layer of a CNN has more than one feature map. If we use multiple feature maps, the kernel tensor-1
becomes four-dimensional: ![]() . Here, width x height is the kernel size,
. Here, width x height is the kernel size, ![]() is the number of input channels, and
is the number of input channels, and ![]() is the number of output feature maps. So, now let's include the number of output feature maps in the preceding formula and update it as follows:
is the number of output feature maps. So, now let's include the number of output feature maps in the preceding formula and update it as follows:
Note: 0<=k<=![]() -1
-1
#########################################################
Numpy examples. To make the discussion above more concrete, lets express the same ideas but in code and with a specific example. Suppose that the input volume is a numpy array X. Then:
- A depth column (or a fibre) at position
(axis_0,axis_1)would be the activationsX[axis_0,axis_1,:]. - A depth slice, or equivalently an activation map at depth
dwould be the activationsX[:,:,d].
Conv Layer Example. Suppose that the input volume X has shape X.shape: (11,11,4). Suppose further that we use no zero padding (P=0), that the filter size is F=5x5, and that the stride is S=2. The output volume would therefore have spatial size (11-5)/2+1 = 4, giving a volume with width and height of 4. The activation map in the output volume (call it V), would then look as follows (only some of the elements are computed in this example): W.shape(5,5,C_in=4, C_out)
V[0,0,0] = np.sum(X[:5,:5,:] * W0) + b0(example of going alongaxis_0)V[1,0,0] = np.sum(X[2:7,:5,:] *W0) + b0V[2,0,0] = np.sum(X[4:9,:5,:] *W0) + b0V[3,0,0] = np.sum(X[6:11,:5,:]*W0) + b0
1 bias per neurons or output chanels C_outnote np.sum(X[:5,:5,:] * W0) ==> sum(X[:5,:5,0]*W0)+sum(X[:5,:5,1]*W0)+sum(X[:5,:5,4-1]*W0)
Remember that in numpy, the operation * above denotes elementwise multiplication between the arrays. Notice also that the weight vector W0 is the weight vector of that neuron and b0 is the bias. Here, W0 is assumed to be of shape W0.shape: (5,5,4), since the filter size is 5 and the depth of the input volume is 4. Notice that at each point, we are computing the dot product as seen before in ordinary neural networks. Also, we see that we are using the same weight and bias (due to parameter sharing), and where the dimensions along the width are increasing in steps of 2 (i.e. the stride).
To construct a second activation map in the output volume, we would have:
V[0,0,1] = np.sum(X[:5,:5,:] * W1) + b1V[1,0,1] = np.sum(X[2:7,:5,:] * W1) + b1V[2,0,1] = np.sum(X[4:9,:5,:] * W1) + b1V[3,0,1] = np.sum(X[6:11,:5,:] * W1) + b1V[0,1,1] = np.sum(X[:5,2:7,:] * W1) + b1(example of going alongaxis_1)V[2,3,1] = np.sum(X[4:9,6:11,:] * W1) + b1( or along both(axis_0, axis_1) )
where we see that we are indexing into the second depth dimension in V (at index 1) because we are computing the second activation map, and that a different set of parameters (W1) is now used. In the example above, we are for brevity leaving out some of the other operations the Conv Layer would perform to fill the other parts of the output array V. Additionally, recall that these activation maps are often followed elementwise through an activation function such as ReLU, but this is not shown here.
#########################################################
# Implementing a CNN in TensorFlow low-level API by using tensorflow.compat.v1
import tensorflow.compat.v1 as tf
import numpy as np
## wrapper functions
def conv_layer(input_tensor, # Tensor("Placeholder:0", shape=(None, 28, 28, 1), dtype=float32)
name, # name='convtest'
kernel_size, # kernel_size=(3, 3)
n_output_channels, # n_output_channels=32
padding_mode='SAME', strides=(1, 1, 1, 1)):
with tf.variable_scope(name):# convtest/
## get n_input_channels:
## input tensor shape:
## [batch x width x height x channels_in]
input_shape = input_tensor.get_shape().as_list() #return [None, 28, 28, 1]
n_input_channels = input_shape[-1]
# [ width, height, C_in, C_out ]
weights_shape = (list(kernel_size) + [n_input_channels, n_output_channels])
weights = tf.get_variable(name='_weights', shape=weights_shape)
print(weights) #
################################################
conv = tf.nn.conv2d(input=input_tensor,
filter=weights,
strides=strides,
padding=padding_mode)
print(conv) #Tensor("convtest/Conv2D:0", shape=(None, 28, 28, 32), dtype=float32)
################################################
biases = tf.get_variable(name='_biases',
initializer=tf.zeros(
shape=[n_output_channels]
)
)
print(biases) #
conv = tf.nn.bias_add(conv, biases,
name='net_pre-activation')
print(conv)#Tensor("convtest/net_pre-activation:0", shape=(None,28,28,32), dtype=float32)
#################################################
conv = tf.nn.relu(conv, name='activation')
print(conv)# Tensor("convtest/activation:0", shape=(None, 28, 28, 32), dtype=float32)
return conv
## testing
g = tf.Graph()
with g.as_default():
x = tf.placeholder(tf.float32, shape=[None, 28, 28, 1])
conv_layer(x, name='convtest', kernel_size=(3, 3), n_output_channels=32)
del g, x To conclude our discussion of computing convolutions in the context of neural networks, let's look at the example in the following figure that shows a convolutional layer, followed by a pooling layer.
In this example, there are three input channels. The kernel tensor is four dimensional ![]() . Each kernel matrix is denoted as
. Each kernel matrix is denoted as ![]() , and there are three of them
, and there are three of them![]() , one for each input channel. Furthermore, there are five such kernels
, one for each input channel. Furthermore, there are five such kernels![]() , accounting for five output feature maps. Finally, there is a pooling layer for subsampling the feature maps, as shown in the following figure:
, accounting for five output feature maps. Finally, there is a pooling layer for subsampling the feature maps, as shown in the following figure:
How many trainable parameters exist in the preceding example?
To illustrate the advantages of convolution, parameter-sharing and sparse connectivity, let's work through an example. The convolutional layer in the network shown in the preceding figure is a four-dimensional tensor. So, there are ![]() parameters associated with the kernel. Furthermore, there is a bias vector for each output feature map of the convolutional layer. Thus, the size of the bias vector is 5
parameters associated with the kernel. Furthermore, there is a bias vector for each output feature map of the convolutional layer. Thus, the size of the bias vector is 5 ![]() . Pooling layers do not have any (trainable) parameters; therefore, we can write the following:
. Pooling layers do not have any (trainable) parameters; therefore, we can write the following:![]() (+5 since the bias vector)
(+5 since the bias vector)
If input tensor is of size ![]() , assuming that the convolution is performed with mode='same', then the output feature maps would be of size
, assuming that the convolution is performed with mode='same', then the output feature maps would be of size ![]() .
.
Note that this number is much smaller than the case if we wanted to have a fully connected layer instead of the convolution layer. In the case of a fully connected layer, the number of parameters for the weight matrix to reach the same number of output units would have been as follows:![]()
In addition, the size of the bias vector is ![]() (one bias element for each output unit). Given that
(one bias element for each output unit). Given that ![]() and
and ![]() , we can see that the difference in the number of trainable parameters is huge.
, we can see that the difference in the number of trainable parameters is huge.
Lastly, as was already mentioned, typically, the convolution operations are carried out by treating an input image with multiple color channels as a stack of matrices; that is, we perform the convolution on each matrix separately and then add the
results, as was illustrated in the previous figure. However, convolutions can also be extended to 3D volumes if you are working with 3D datasets, for example, as shown in the paper VoxNet: A 3D Convolutional Neural Network for Real-Time Object
Recognition (2015), by Daniel Maturana and Sebastian Scherer, which can be accessed at https://www.ri.cmu.edu/pub_files/2015/9/voxnet_maturana_scherer_iros15.pdf.
In the next section, we will talk about how to regularize a neural network.
Regularizing a neural network with dropout
Choosing the size of a network, whether we are dealing with a traditional (fully connected) neural network or a CNN, has always been a challenging problem. For instance, the size of a weight matrix and the number of layers need to be tuned to
achieve a reasonably good performance.
You will recall from Chapter 14, Going Deeper – The Mechanics of TensorFlow, thata simple network without any hidden layer could only capture a linear decision boundary, which is not sufficient for dealing with an exclusive or (XOR) or similar
problem.The capacity of a network refers to the level of complexity of the function that it can learn. Small networks, networks with a relatively small number of parameters, have a low capacity and are therefore likely to be under fit, resulting in poor performance since they cannot learn the underlying structure of complex datasets.
However, very large networks may more easily result in overfitting, where the network will memorize the training data and do extremely well on the training set while achieving poor performance on the held-out test set. When we deal with real-world machine learning problems, we do not know how large the network should be a priori.
One way to address this problem is to build a network with a relatively large capacity (in practice, we want to choose a capacity that is slightly larger than necessary) to do well on the training set. Then, to prevent overfitting, we can apply
one or multiple regularization schemes to achieve good generalization performance on new data, such as the held-out test set. A popular choice for regularization is L2 regularization(to minimize the sum of the unpenalized cost function plus the penalty term ![]() ), which we discussed previously in cp4 Training Sets Preprocessing_StringIO_dropna_categorical_feature_Encode_Scale_L1_L2_bbox_to_anchor
), which we discussed previously in cp4 Training Sets Preprocessing_StringIO_dropna_categorical_feature_Encode_Scale_L1_L2_bbox_to_anchor
https://blog.csdn.net/Linli522362242/article/details/108230328.
In Chapter 3, A Tour of Machine Learning Classifiers Using scikit-learn, we covered L1 and L2 regularization. In the section Tackling overfitting via regularization, you saw that both techniques, L1 and L2 regularization, can prevent or reduce the effect
of overfitting by adding a penalty to the loss that results in shrinking the weight parameters during training. While both L1 and L2 regularization can be used for NNs as well, with L2 being the more common choice of the two, there are other methods for regularizing NNs, such as dropout, which we discuss in this section. But before we move on to discussing dropout, to use L2 regularization within a convolutional or fully connected (dense) network, you can simply add the L2 penalty
to the loss function by setting the kernel_regularizer of a particular layer when using the Keras API, as follows (it will then automatically modify the loss function accordingly)
from tensorflow import keras
conv_layer = keras.layers.Conv2D(filters=16,
kernel_size=(3,3),
kernel_regularizer=keras.regularizers.l2(0.001)
)
fc_layer = keras.layers.Dense(units=16,
kernel_regularizer=keras.regularizers.l2(0.001)
) In recent years, another popular regularization technique called dropout has emerged that works amazingly well for regularizing (deep) neural networks (Dropout: a simple way to prevent neural networks from overfitting, Nitish
Srivastava and. others, Journal of Machine Learning Research 15.1, pages 1929-1958, 2014, http://www.jmlr.org/papers/volume15/srivastava14a/srivastava14a.pdf).
Intuitively, dropout can be considered as the consensus (averaging) of an ensemble of models. In ensemble learning, we train several models independently. During prediction, we then use the consensus of all the trained models. However, both
training several models and collecting and averaging the output of multiple models is computationally expensive. Here, dropout offers a workaround with an efficient way to train many models at once and compute their average predictions at test or prediction time.
Dropout is usually applied to the hidden units of higher layers. During the training phase of a neural network, a fraction of the hidden units is randomly dropped at every iteration with probability ![]() (or the keep probability
(or the keep probability ![]() ). This dropout probability is determined by the user and the common choice is
). This dropout probability is determined by the user and the common choice is ![]() , as discussed in the previously mentioned article by Nitish Srivastava and others, 2014. When dropping a certain fraction of input neurons, the weights associated with the remaining neurons are rescaled to account for the missing (dropped) neurons. ###The hyperparameter p is called the dropout rate, and it is typically set between 10% and 50%: closer to 20–30% in recurrent neural nets, and closer to 40–50% in convolutional neural networks###
, as discussed in the previously mentioned article by Nitish Srivastava and others, 2014. When dropping a certain fraction of input neurons, the weights associated with the remaining neurons are rescaled to account for the missing (dropped) neurons. ###The hyperparameter p is called the dropout rate, and it is typically set between 10% and 50%: closer to 20–30% in recurrent neural nets, and closer to 40–50% in convolutional neural networks###
The effect of this random dropout forces the network to learn a redundant representation of the data. Therefore, the network cannot rely on an activation of any set of hidden units since they may be turned off at any time during training and is forced to learn more general and robust patterns from the data.
This random dropout can effectively prevent overfitting. The following figure shows an example of applying dropout with probability ![]() during the training phase, thereby half of the neurons become inactive randomly. However, during prediction, all neurons will contribute to computing the pre-activations of the next layer.
during the training phase, thereby half of the neurons become inactive randomly. However, during prediction, all neurons will contribute to computing the pre-activations of the next layer.
As shown here, one important point to remember is that units may drop randomly during training only, while for the evaluation phase, all the hidden units must be active (for instance, ![]() or
or ![]() ). To ensure that the overall activations are on the same scale during training and prediction, the activations of the active neurons have to be scaled appropriately (for example, by halving the activation if the dropout probability was set to
). To ensure that the overall activations are on the same scale during training and prediction, the activations of the active neurons have to be scaled appropriately (for example, by halving the activation if the dropout probability was set to ![]() ).
).
However, since it is inconvenient to always scale activations when we make predictions in practice, TensorFlow and other tools scale the activations during training (for example, by doubling the activations if the dropout probability was set to ![]() ).This approach is commonly referred to as inverse dropout.
).This approach is commonly referred to as inverse dropout.
As mentioned previously, the relationship between model ensembles and dropout is not immediately obvious. However, consider that in dropout, we have a different model for each mini-batch (due to setting the weights to zero randomly during each forward pass).
Then, via iterating over the mini-batches, we essentially sample over ![]() models, where h is the number of hidden units( these hidden units may dropped OR not ==> 2).
models, where h is the number of hidden units( these hidden units may dropped OR not ==> 2).
The restriction and aspect that distinguishes dropout from regular ensembling, however, is that we share the weights over these "different models", which can be seen as a form of regularization. Then, during "inference" (for instance, predicting
the labels in the test dataset), we can average over all these different models that we sampled over during training. This is very expensive, though.
Then, averaging the models, that is, computing the geometric mean of the classmembership probability that is returned by a model, i, can be computed as follows:
Now, the trick behind dropout is that this geometric mean of the model ensembles (here, ![]() models) can be approximated by scaling the predictions of the last (or final) model sampled during training by a factor of 1/(1 – p) ### the dropout probability is p###, which is much cheaper than computing the geometric mean explicitly using the previous equation. (In fact, the approximation is exactly equivalent to the true geometric mean if we consider linear models.)
models) can be approximated by scaling the predictions of the last (or final) model sampled during training by a factor of 1/(1 – p) ### the dropout probability is p###, which is much cheaper than computing the geometric mean explicitly using the previous equation. (In fact, the approximation is exactly equivalent to the true geometric mean if we consider linear models.)
So, what is the relationship between dropout and ensemble learning? Since we drop different hidden neurons at each iteration, effectively we are training different models. When all these models are finally trained, we set the keep probability to 1 and use all the hidden units. This means we are taking the average activation from all the hidden units.
Loss functions for classification
In Chapter 13, Parallelizing Neural Network Training with TensorFlow, we saw different activation functions, such as ReLU, sigmoid, and tanh. Some of these activation functions, like ReLU, are mainly used in the intermediate (hidden) layers of an NN to add non-linearities to our model. But others, like sigmoid (for binary) and softmax (for multiclass), are added at the last (output) layer, which results in classmembership probabilities as the output of the model. If the sigmoid or softmax activations are not included at the output layer, then the model will compute the logits instead of the class-membership probabilities.
Focusing on classification problems here, depending on the type of problem (binary versus multiclass) and the type of output (logits versus probabilities), we should choose the appropriate loss function to train our model. Binary cross-entropy is the loss function for a binary classification (with a single output unit), and categorical cross-entropy is the loss function for multiclass classification. In the Keras API, two options for categorical cross-entropy loss are provided, depending on whether the ground truth labels are in a one-hot encoded format (for example, [0, 0, 1, 0]), or provided as integer labels (for example, y=2), which is also known as "sparse" representation in the context of Keras.
The following table describes three loss functions available in Keras for dealing with all three cases: binary classification, multiclass with one-hot encoded ground truth labels, and multiclass with integer (sparse) labels. Each one of these three loss
functions also has the option to receive the predictions in the form of logits or classmembership probabilities:
https://blog.csdn.net/Linli522362242/article/details/96480059  =np.log( 0.69/(1-0.69) )=0.8
=np.log( 0.69/(1-0.69) )=0.8
-
BinaryCrossentropy() #https://blog.csdn.net/Linli522362242/article/details/96480059
sigmoid: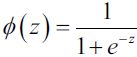
-
CategoricalCrossentropy() #https://blog.csdn.net/Linli522362242/article/details/104124771
logits =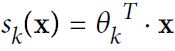 :
: 
Softmax function:
from_logits=Falsefrom_logits=True#Cross entropy cost function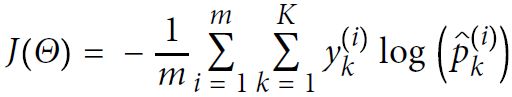
-
SparseCategoricalCrossentropy()from_logits=Falsefrom_logits=True
import tensorflow_datasets as tfds
############## Binary Crossentropy loss ##############
bce_probas = tf.keras.losses.BinaryCrossentropy( from_logits=False )#classmembership probabilities
bce_logits = tf.keras.losses.BinaryCrossentropy( from_logits=True )
logits = tf.constant([0.8]) # z=w^T *X
probas = tf.keras.activations.sigmoid(logits) # 1/(1+np.exp(-0.8))
# probas :
#
tf.print(
'BinaryCrossEntropy (w Probas): {:.4f}'.format(
bce_probas( y_true=[1], y_pred=probas )
),
' # BinaryCrossEntropy (w Logits): {:.4f}'.format( # round(-np.log(0.69),4)=0.3711
bce_logits( y_true=[1], y_pred=logits) #if y=1, -log( sigmoid(z))
) #if y=0, -log(1-sigmoid(z))
)
############## Categorical Crossentropy loss ##############
cce_probas = tf.keras.losses.CategoricalCrossentropy( from_logits=False )
cce_logits = tf.keras.losses.CategoricalCrossentropy( from_logits=True )
logits = tf.constant([ [1.5, 0.8, 2.1] ])
probas = tf.keras.activations.softmax( logits )
# probas
#
# np.exp(1.5) / (np.exp(1.5) + np.exp(0.8) + np.exp(2.1)) : 0.3013224
# np.exp(0.8) / (np.exp(1.5) + np.exp(0.8) + np.exp(2.1)) : 0.1496323
# np.exp(2.1) / (np.exp(1.5) + np.exp(0.8) + np.exp(2.1)) : 0.5490452
tf.print(
'CategoricalCrossentropy (w Probas): {:.4f}'.format(
#set the position on np.argmax( logits )=1, the rest is set to 0
cce_probas(y_true=[0,0,1], y_pred=probas)
),
' # CategoricalCrossentropy (w Logits): {:.4f}'.format(
#set the position on np.argmax( logits )=1, the rest is set to 0
cce_logits(y_true=[0,0,1], y_pred=logits)
)# -1 * np.log(np.exp(2.1) / (np.exp(1.5) + np.exp(0.8) + np.exp(2.1)))
# =0.5996
#OR
# -tf.reduce_sum ( [0,0,1] * np.log(probas) ,keepdims=True) # for 1 instance
# tf.Tensor: shape=(1, 1), dtype=float64, numpy=array([[0.59957439]])>
)
############## Sparse Categorical Crossentropy ##############
sp_cce_probas = tf.keras.losses.SparseCategoricalCrossentropy( from_logits=False )
sp_cce_logits = tf.keras.losses.SparseCategoricalCrossentropy( from_logits=True )
tf.print(
'SparseCategoricalCrossentropy (w Probas): {:.4f}'.format(
sp_cce_probas(y_true=[2], y_pred=probas)),
'# SparseCategoricalCrossentropy(w Logits): {:.4f}'.format(
sp_cce_logits(y_true=[2], y_pred=logits))
)# -np.log(probas)[:,np.argmax( logits )] # for 1 instance
# array([0.5995744], dtype=float32) Note that sometimes, you may come across an implementation where a categorical cross-entropy loss is used for binary classification. Typically, when we have a binary classification task, the model returns a single output value for each example.
We interpret this single model output as the probability of the positive class (for example, class 1), P[class = 1]. In a binary classification problem, it is implied that P[class = 0] = 1 – P[class = 1]; hence, we do not need a second output unit in order to obtain the probability of the negative class. However, sometimes practitioners choose to return two outputs for each training example and interpret them as probabilities of each class: P[class = 0] versus P[class = 1]. Then, in such a case, using a softmax function (instead of the logistic sigmoid) to normalize the outputs (so that they sum
to 1) is recommended, and categorical cross-entropy is the appropriate loss function.
Please note that computing the cross-entropy loss by providing the logits, and not the class-membership probabilities, is usually preferred due to numerical stability reasons. If we provide logits as inputs to the loss function and set from_logits=True, the respective TensorFlow function uses a more efficient implementation to compute the loss and derivative of the loss with respect to the weights. This is possible since certain mathematical terms cancel and thus don't have to be computed explicitly when providing logits as inputs.
Implementing a deep convolutional neural network using TensorFlow
In Chapter 13, Parallelizing Neural Network Training with TensorFlow, you may recall that we implemented a multilayer neural network for handwritten digit recognition problems, using different API levels of TensorFlow. You may also recall that we achieved about 97 percent accuracy.
So now, we want to implement a CNN to solve this same problem and see its predictive power in classifying handwritten digits. Note that the fully connected layers that we saw in the Chapter 13, Parallelizing Neural Network Training with
TensorFlow were able to perform well on this problem. However, in some applications, such as reading bank account numbers from handwritten digits, even tiny mistakes can be very costly. Therefore, it is crucial to reduce this error as much as possible.
The multilayer CNN architecture
The architecture of the network that we are going to implement is shown in the following figure. The input is 28 x 28 grayscale images. Considering the number of channels (which is 1 for grayscale images) and a batch of input images, the input tensor's dimensions will be batchsize x 28 x 28 x 1.
The input data goes through two convolutional layers that have a kernel size of 5 x 5. The first convolution has 32 output feature maps, and the second one has 64 output feature maps. Each convolution layer is followed by a subsampling layer in the form of a max-pooling operation.
Then a fully-connected layer passes the output to a second fully-connected layer, which acts as the final softmax output layer. The architecture of the network that we are going to implement is shown in the following figure:
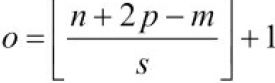
The dimensions of the tensors in each layer are as follows: ( kernel_size=(5, 5), pool_size=(2,2) )
- Input:
 ==>input n=28, Assuming padding='same' then P=2, stride s=1,
==>input n=28, Assuming padding='same' then P=2, stride s=1,
output shape=(28+2*2-5)/1+1=28 - Conv_1:[

 ] ==> filters=32 feature maps,
] ==> filters=32 feature maps, - Pooling_1:[

 ] and pool_size=(2,2) ==> strides=(2,2), padding='valid' and p=0,
] and pool_size=(2,2) ==> strides=(2,2), padding='valid' and p=0,
output shape=(28+2*0-2)/2+1=14 - Conv_2:[

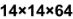 ] ==> input n=14, Assuming padding='same' then P=2, stride s=2,
] ==> input n=14, Assuming padding='same' then P=2, stride s=2,
output shape=(14+2*2-5)/2+1=7 - Pooling_2:[

 ]==>filters=64 feature maps,
]==>filters=64 feature maps, - FC_1:[
 1024]
1024] - FC_2 and softmax layer:[
 10]
10]
For the convolutional kernels, we are using strides=1 such that the input dimensions are preserved in the resulting feature maps. For the pooling layers, we are using strides=2 to subsample the image and shrink the size of the output
feature maps. We will implement this network using the TensorFlow Keras API.
# Implementing a CNN in TensorFlow low-level API by using tensorflow.compat.v1
import tensorflow.compat.v1 as tf
def fc_layer(input_tensor, # Tensor("Placeholder:0", shape=(None, 7, 7, 64), dtype=float32)
name, # name='fctest'
n_output_units, # n_output_units=1024
activation_fn=None):# activation_fn=tf.nn.relu
with tf.variable_scope(name):
input_shape = input_tensor.get_shape().as_list()[1:] # return [7, 7, 64]
n_input_units = np.prod(input_shape) # return 3136 = 7*7*64
if len(input_shape) > 1:
input_tensor = tf.reshape(input_tensor,
shape=(-1, n_input_units)
)#Tensor("fctest/Reshape:0", shape=(None,3136), dtype=float32)
weights_shape = [n_input_units, n_output_units] #[3136, 1024]
weights = tf.get_variable(name='_weights',
shape=weights_shape)
print(weights) #
biases = tf.get_variable(name='_biases',
initializer=tf.zeros(
shape=[n_output_units]
)
)
print(biases) #
# mat_shape(batches, 3136) x mat_shape(3136, 1024) ==> mat_shape(batches, 1024)
layer = tf.matmul(input_tensor, weights)#shape(None, 1024)
print(layer) # Tensor("fctest/MatMul:0", shape=(None, 1024), dtype=float32)
layer = tf.nn.bias_add(layer, biases,
name='net_pre-activation')
print(layer) # Tensor("fctest/net_pre-activation:0", shape=(None, 1024), dtype=float32)
if activation_fn is None:
return layer
layer = activation_fn(layer, name='activation')
print(layer) # Tensor("fctest/activation:0", shape=(None, 1024), dtype=float32)
return layer
## testing:
g = tf.Graph()
with g.as_default():
x = tf.placeholder(tf.float32, shape=[None, 7, 7, 64])
fc_layer(x, name='fctest', n_output_units=1024,
activation_fn=tf.nn.relu)
del g, x Loading and preprocessing the data
You will recall that in Chapter 13, Parallelizing Neural Network Training with TensorFlow, you learned two ways of loading available datasets from the tensorflow_datasets module. One approach is based on a three-step process, and a simpler method uses a function called load, which wraps those three steps. Here, we will use the first method. The three steps for loading the MNIST dataset are as follows:
import tensorflow_datasets as tfds
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
print( tfds.list_builders() )## MNIST dataset
mnist_bldr = tfds.builder('mnist')
mnist_bldr.download_and_prepare()# Download the data, prepare it, and write it to disk
datasets = mnist_bldr.as_dataset( shuffle_files=False )# Load data from disk as tf.data.Datasets
print( datasets.keys() )
mnist_train_orig, mnist_test_orig = datasets['train'], datasets['test'] #'PrefetchDataset' object![]()
The MNIST dataset comes with a pre-specified training and test dataset partitioning scheme, but we also want to create a validation split from the train partition. Notice that in the third step, we used an optional argument, shuffle_files=False, in the
.as_dataset() method. This prevented initial shuffling, which is necessary for us since we want to split the training dataset into two parts: a smaller training dataset and a validation dataset. (Note: if the initial shuffling was not turned off, it would incur reshuffling of the dataset every time we fetched a mini-batch of data.
An example of this behavior is shown in the online contents of this chapter, where you can see that the number of labels in the validation datasets changes due to reshuffling of the train/validation splits. This can cause false performance estimation
of the model, since the train/validation datasets are indeed mixed.) We can split thetrain/validation datasets as follows:
BUFFER_SIZE = 10000
BATCH_SIZE = 64
NUM_EPOCHS = 20
mnist_train = mnist_train_orig.map(
lambda item: (tf.cast( item['image'], tf.float32 )/255.0,
tf.cast( item['label'], tf.int32 )
)
)
mnist_test = mnist_test_orig.map(
lambda item: (tf.cast( item['image'], tf.float32 )/255.0,
tf.cast( item['label'], tf.int32)
)
)
tf.random.set_seed(1)
mnist_train = mnist_train.shuffle( buffer_size=BUFFER_SIZE,
reshuffle_each_iteration=False
) # ==> preprocess_map ==> batch
mnist_valid = mnist_train.take(10000).batch(BATCH_SIZE)
mnist_train = mnist_train.skip(10000).batch(BATCH_SIZE)Now, after preparing the dataset, we are ready to implement the CNN we just described.
Implementing a CNN using the TensorFlow Keras API
For implementing a CNN in TensorFlow, we use the Keras Sequential class to stack different layers, such as convolution, pooling, and dropout, as well as the fully connected (dense) layers. The Keras layers API provides classes for each one:
tf.keras.layers.Conv2D for a two-dimensional convolution layer; tf.keras.layers.MaxPool2D and tf.keras.layers.AvgPool2D for subsampling (maxpooling and average-pooling); and tf.keras.layers.Dropout for regularization using dropout. We will go over each of these classes in more detail.
Configuring CNN layers in Keras
Constructing a layer with the Conv2D class requires us to specify the number of output filters (which is equivalent to the number of output feature maps) and kernel sizes.
In addition, there are optional parameters that we can use to configure a convolutional layer. The most commonly used ones are the strides (with a default value of 1 in both x, y dimensions) and padding, which could be same or valid. Additional configuration parameters are listed in the official documentation: https://www.tensorflow.org/versions/r2.0/api_docs/python/tf/keras/layers/Conv2D.
It is worth mentioning that usually, when we read an image, the default dimension for the channels is the last dimension of the tensor array. This is called the "NHWC" format, where N stands for the number of images within the batch, H and W stand for height and width, respectively, and C stands for channels.
Note that the Conv2D class assumes that inputs are in the NHWC format by default. (Other tools, such as PyTorch, use an NCHW format.) However, if you come across some data whose channels are placed at the first dimension (the first dimension after the batch dimension, or second dimension considering the batch dimension), you would need to swap the axes in your data to move the channels to the last dimension. Or, an alternative way to work with an NCHW-formatted input is to set data_format="channels_first". After the layer is constructed, it can be called by providing a four-dimensional tensor, with the first dimension reserved for a batch of examples; depending on the data_format argument, either the second or the fourth dimension corresponds to the channel; and the other two dimensions are the spatial dimensions.
As shown in the architecture of the CNN model that we want to build, each convolution layer is followed by a pooling layer for subsampling (reducing the size of feature maps). The MaxPool2D and AvgPool2D classes construct the max-pooling and average-pooling layers, respectively. The argument pool_size determines the size of the window (or neighborhood) that will be used to compute the max or mean operations. Furthermore, the strides parameter can be used to configure the pooling layer, as we discussed earlier.
Finally, the Dropout class will construct the dropout layer for regularization, with the argument rate used to determine the probability of dropping the input units during the training. When calling this layer, its behavior can be controlled via an argument named training, to specify whether this call will be made during training or during the inference.
Configuring CNN layers in Keras
- Conv2D:
tf.keras.layers.Conv2Dfilterskernel_sizestridespadding
- MaxPool2D:
tf.keras.layers.MaxPool2Dpool_sizestridespadding
- Dropout
tf.keras.layers.Dropout2Drate
Constructing a CNN in Keras
Now that you have learned about these classes, we can construct the CNN model that was shown in the previous figure. In the following code, we will use the Sequential class and add the convolution and pooling layers:
model = tf.keras.Sequential()
model.add(tf.keras.layers.Conv2D(
filters=32, kernel_size=(5,5),
strides=(1,1), padding='same', #==> input_shape==output_shape
data_format='channels_last',
name='conv_1', activation='relu'
))# output_shape =(n+2p-m)/s +1 =(28+2*2-5)/1 +1=28 and p=2
model.add(tf.keras.layers.MaxPool2D(
pool_size=(2,2), # strides: If None, it will default to `pool_size`.
name='pool_1' # padding='valid' default
))# output_shape =(n+2p-m)/s +1 =(28+2*0-2)/2 +1=14 and p=0
model.add(tf.keras.layers.Conv2D(
filters=64, kernel_size=(5,5),
strides=(1,1), padding='same', #==> input_shape==output_shape
name='conv_2', activation='relu'
))# output_shape =(n+2p-m)/s +1 =(14+2*2-5)/1 +1=14 and p=2
model.add(tf.keras.layers.MaxPool2D(
pool_size=(2,2),
name='pool_2'
))# output_shape =(n+2p-m)/s +1 =(14+2*0-2)/2 +1=7 and p=0 So far, we have added two convolution layers to the model. For each convolutional layer, we used a kernel of size 5 × 5 and 'same' padding. As discussed earlier, using padding='same' preserves the spatial dimensions (vertical and horizontal
dimensions) of the feature maps such that the inputs and outputs have the same height and width (and the number of channels may only differ in terms of the number of filters used). The max-pooling layers with pooling size 2 × 2 and strides of 2 will reduce the spatial dimensions by half. (Note that if the strides parameter is not specified in MaxPool2D, by default, it is set equal to the pooling size.)
While we can calculate the size of the feature maps at this stage manually, the Keras API provides a convenient method to compute this for us:
model.compute_output_shape( input_shape=(16,28,28,1))![]()
By providing the input shape as a tuple specified in this example, the method compute_output_shape calculated the output to have a shape (16, 7, 7, 64), indicating feature maps with 64 channels and a spatial size of 7 × 7 . The first dimension corresponds to the batch dimension, for which we used 16 arbitrarily. We could have used None instead, that is, input_shape=(None, 28, 28, 1).
The next layer that we want to add is a dense (or fully connected) layer for implementing a classifier on top of our convolutional and pooling layers. The input to this layer must have rank 2, that is, shape ![]() . Thus, we need to flatten the output of the previous layers to meet this requirement for the dense layer:
. Thus, we need to flatten the output of the previous layers to meet this requirement for the dense layer:
model.add(tf.keras.layers.Flatten()) #2D##############
model.compute_output_shape( input_shape=(16,28,28,1) ) # 7x7* 64 feature maps=3136![]()
As the result of compute_output_shape indicates, the input dimensions for the dense layer are correctly set up. Next, we will add two dense layers with a dropout layer in between:
model.add(tf.keras.layers.Dense(
units=1024, name='fc_1',
activation='relu'
))
model.add(tf.keras.layers.Dropout(
rate=0.5
))
model.add(tf.keras.layers.Dense(
units=10, name='fc_2', # units=10 since 10 classes or labels
activation='softmax'
))The last fully connected layer, named 'fc_2', has 10 output units for the 10 class labels in the MNIST dataset. Also, we use the softmax activation to obtain the classmembership probabilities of each input example, assuming that the classes are mutually exclusive, so the probabilities for each example sum to 1. (This means that a training example can belong to only one class.) Based on what we discussed in the section Loss functions for classification, which loss should we use here? Remember that for a multiclass classification with integer (sparse) labels (as opposed to one-hot encoded labels), we use SparseCategoricalCrossentropy. The following code will call the build() method for late variable creation and compile the model:
tf.random.set_seed(1)
model.build(input_shape=(None, 28,28,1))
model.compute_output_shape(input_shape=(16,28,28,1))![]()
model.summary()model.compile(optimizer=tf.keras.optimizers.Adam(),
loss=tf.keras.losses.SparseCategoricalCrossentropy(),
metrics=['accuracy'])# same as `tf.keras.metrics.SparseCategoricalAccuracy(name='accuracy')
The Adam optimizer
Note that in this implementation, we used the tf.keras.optimizers.Adam() class for training the CNN model. The
Adam optimizer is a robust, gradient-based optimization method suited to nonconvex optimization and machine learning
problems. Two popular optimization methods inspired Adam: RMSProp and AdaGrad. The key advantage of Adam is in the choice of update step size derived from the running average of gradient moments. Please feel free to read more about the Adam optimizer in the manuscript, Adam: A Method for Stochastic Optimization, Diederik P. Kingma and Jimmy Lei Ba, 2014. The article is freely available at https://arxiv.org/abs/1412.6980.
11_Training Deep Neural Networks_3_Adam_Learning Rate Scheduling_Decay_np.argmax(」)_lambda语句_Regular: https://blog.csdn.net/Linli522362242/article/details/107086444
As you already know, we can train the model by calling the fit() method. Note that using the designated methods for training and evaluation (like evaluate() and predict()) will automatically set the mode for the dropout layer and rescale the hidden units appropriately so that we do not have to worry about that at all. Next, we will train this CNN model and use the validation dataset that we created for monitoring the learning progress:
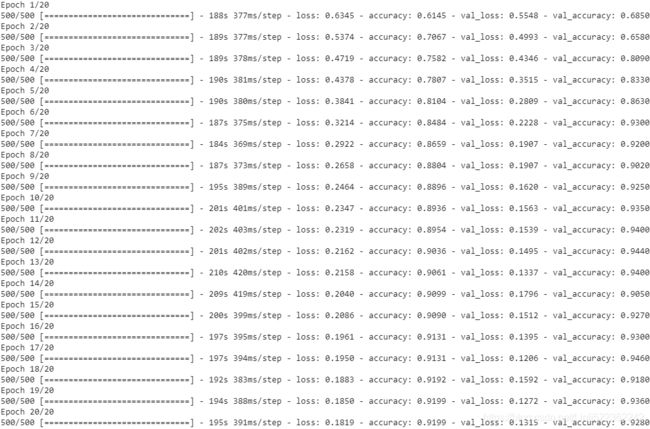
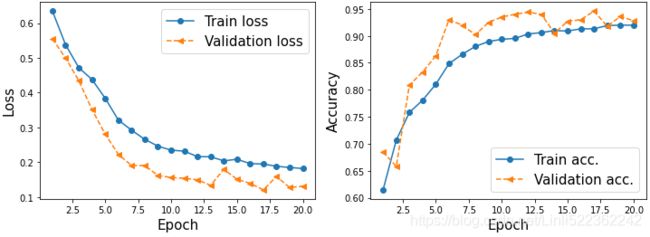
history = model.fit( mnist_train, epochs=NUM_EPOCHS, validation_data=mnist_valid, # Xs,y_label
shuffle=True)Once the 20 epochs of training are finished, we can visualize the learning curves:
import matplotlib.pyplot as plt
import numpy as np
hist = history.history
# hist.keys() # dict_keys(['loss', 'accuracy', 'val_loss', 'val_accuracy'])
x_arr = np.arange( len(hist['loss']) ) + 1
fig = plt.figure( figsize=(12,4) )
ax = fig.add_subplot(1,2,1)
ax.plot( x_arr, hist['loss'], '-o', label='Train loss')
ax.plot( x_arr, hist['val_loss'], '--<', label='Validation loss')
ax.set_xlabel('Epoch', size=15)
ax.set_ylabel('Loss', size=15)
ax.legend(fontsize=15)
ax = fig.add_subplot(1,2,2)
ax.plot( x_arr, hist['accuracy'], '-o', label='Train acc.' )
ax.plot( x_arr, hist['val_accuracy'], '--<', label='Validation acc.' )
ax.set_xlabel('Epoch', size=15)
ax.set_ylabel('Accuracy', size=15)
ax.legend(fontsize=15)
plt.show()As you already know from the two previous chapters, evaluating the trained model on the test dataset can be done by calling the .evaluate() method:
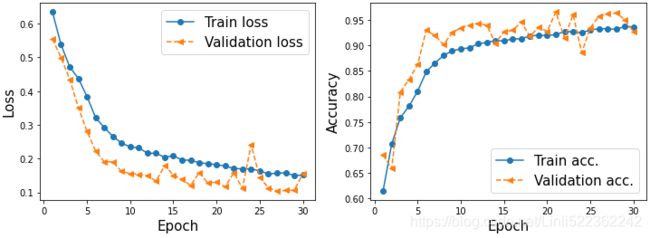
test_results = model.evaluate( mnist_test.batch(20) )
print( 'Test Accuracy: {:.2f}%'.format(test_results[1]*100) )The CNN model achieves an accuracy of 99.32%. Remember that in Chapter 14, Going Deeper – The Mechanics of TensorFlow, we got approximately 90% accuracy using the Estimator DNNClassifier.
Finally, we can get the prediction results in the form of class-membership probabilities and convert them to predicted labels by using the tf.argmax function to find the element with the maximum probability. We will do this for a batch of 12 examples and visualize the input and predicted labels:
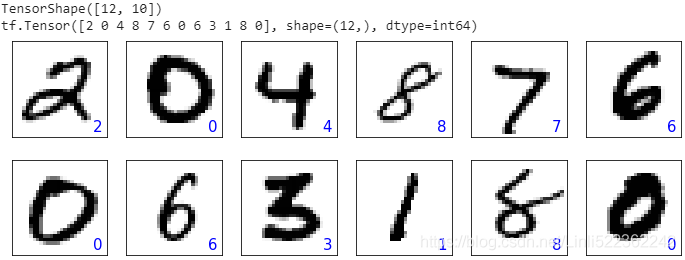
batch_test = next( iter(mnist_test.batch(12)) ) #iter() 函数用来生成迭代器对象
preds = model(batch_test[0])
tf.print( preds.shape ) # TensorShape([12, 10])
preds = tf.argmax( preds, axis=1 )
print(preds)
fig = plt.figure( figsize=(12,4) )
for i in range(12): #12 instances
ax = fig.add_subplot( 2,6, i+1 )
ax.set_xticks([]) #remove ticks
ax.set_yticks([])
img = batch_test[0][i, :, :, 0] # batch_test[0].shape : TensorShape([12, 28, 28, 1])
# gray:0-255 级灰度,0:黑色,1:白色,黑底白字;
# gray_r:翻转 gray 的显示,如果 gray 将图像显示为黑底白字,gray_r 会将其显示为白底黑字;
ax.imshow( img, cmap='gray_r' )
ax.text( 0.9, 0.1, '{}'.format(preds[i]), # the location(0.9, 0.1) of the anchor point
size=15, color='blue',
horizontalalignment='center', # center of text_box(square box) as the anchor point
verticalalignment='center', # center of text_box(square box) as the anchor point
transform=ax.transAxes
)
plt.show()In this set of plotted examples, all the predicted labels are correct.
We leave the task of showing some of the misclassified digits, as we did in Chapter12, Implementing a Multilayer Artificial Neural Network from Scratch, as an exercise for the reader.
import os
if not os.path.exists('models'):
os.mkdir('models')
model.save('models/mnist-cnn.h5')https://colab.research.google.com/drive/1YWrnO1GKoy1wQmjgN1cHGojh5npWj0dG#scrollTo=lTG6COrVvZQz
Gender classification from face images using a CNN
In this section, we are going to implement a CNN for gender classification from face images using the CelebA dataset https://www.tensorflow.org/datasets/catalog/celeb_a. As you already saw in Chapter 13, Parallelizing Neural Network Training with TensorFlow, the CelebA dataset contains 202,599 face images of celebrities. In addition, 40 binary facial attributes are available for each image, including gender (male or female) and age (young or old).
Based on what you have learned so far, the goal of this section is to build and train a CNN model for predicting the gender attribute from these face images. Here, for simplicity, we will only be using a small portion of the training data (16,000
training examples) to speed up the training process. However, in order to improve the generalization performance and reduce overfitting on such a small dataset, we will use a technique called data augmentation数据增强.
Loading the CelebA dataset
First, let's load the data similarly to how we did in the previous section for the MNIST dataset. CelebA data comes in three partitions: a training dataset, a validation dataset, and a test dataset. Next, we will implement a simple function to count the number of examples in each partition:
import tensorflow as tf
import tensorflow_datasets as tfds
import numpy as np
from IPython.display import Image
import matplotlib.pyplot as plt
%matplotlib inline
celeba_bldr = tfds.builder( 'celeb_a' )
celeba_bldr.download_and_prepare() # Download the data, prepare it, and write it to disk
celeba = celeba_bldr.as_dataset( shuffle_files=False) # Load data from disk as tf.data.Datasets
print( celeba.keys() )![]()
celeba_train = celeba['train']
celeba_valid = celeba['validation']
celeba_test = celeba['test']
def count_items(ds):
n = 0
for _ in ds:
n+=1
return n
print('Train set: {}'.format( count_items(celeba_train) ) )
print('Validation: {}'.format( count_items(celeba_valid) ) )
print('Test set: {}'.format( count_items(celeba_test) ) )So, instead of using all the available training and validation data, we will take a subset of 16,000 training examples and 1,000 examples for validation, as follows:
celeba_train = celeba_train.take(16000)
celeba_valid = celeba_valid.take(1000)
print('Train set: {}'.format( count_items(celeba_train) ) )
print('Validation: {}'.format( count_items(celeba_valid) ) )![]()
It is important to note that if the shuffle_files argument in celeba_bldr.as_dataset() was not set to False, we would still see 16,000 examples in the training dataset and 1,000 examples for the validation dataset. However, at each iteration, it would reshuffle the training data and take a new set of 16,000 examples. This would defeat the purpose, as our goal here is to intentionally train our model with a small dataset. Next, we will discuss data augmentation as a technique for boosting the performance of deep NNs.
Image transformation and data augmentation
Data augmentation summarizes a broad set of techniques for dealing with cases where the training data is limited. For instance, certain data augmentation techniques allow us to modify or even artificially synthesize合成 more data and
thereby boost the performance of a machine or deep learning model by reducing overfitting. While data augmentation is not only for image data, there are a set of transformations uniquely applicable to image data, such as cropping parts of an
image, flipping, changing the contrast, brightness, and saturation. Let's see some of these transformations that are available via the tf.image module.
In the following code block, we will first get five examples from the celeba_train dataset and
apply five different types of transformation:
- 1) cropping an image to a bounding box,
- 2) flipping an image horizontally,
- 3) adjusting the contrast,
- 4) adjusting the brightness, and
- 5) center-cropping an image and resizing the resulting image back to its original size, (218, 178).
In the following code, we will visualize the results of these transformations, showing each one in a separate column for comparison:
## take 5 examples:
examples = []
for example in celeba_train.take(5):
examples.append( example['image'])
fig = plt.figure( figsize=(16, 8.5) )
## Column 1: cropping to a bounding-box
ax = fig.add_subplot(2,5,1)
ax.imshow( examples[0] )
ax = fig.add_subplot(2,5,6)
ax.set_title( 'Crop to a \nbounding-box', size=15 )
# tf.image.crop_to_bounding_box(
# image,
# offset_height, # Vertical coordinate of the top-left corner of the result in the input.
# offset_width, # Horizontal coordinate of the top-left corner of the result in the input.
# target_height, # Height of the result.
# target_width # Width of the result.
# )
img_cropped = tf.image.crop_to_bounding_box( examples[0], 50, 20, 128, 128 )
ax.imshow( img_cropped )
## Column 2: flipping (horizontally)
ax = fig.add_subplot(2,5,2)
ax.imshow(examples[1])
ax = fig.add_subplot(2,5,7)
ax.set_title( 'Flip (horizontal)', size=15 )
img_flipped = tf.image.flip_left_right(examples[1])
ax.imshow( img_flipped )
## Column 3: adjust constrast
ax = fig.add_subplot(2,5,3)
ax.imshow(examples[2])
ax = fig.add_subplot(2,5,8)
ax.set_title('Adjust constrast', size=15)
# For each channel, this Op computes the mean of the image pixels in the channel and
# then adjusts each component x of each pixel to (x-mean) *contrast_factor +mean.
img_adj_constrast = tf.image.adjust_contrast( examples[2], contrast_factor=2)
ax.imshow( img_adj_constrast )
## Column 4: adjust brightness
ax = fig.add_subplot( 2,5,4 )
ax.imshow( examples[3] )
ax = fig.add_subplot( 2,5,9 )
ax.set_title('Adjust brightness', size=15 ) # *1.3
img_adj_brightness = tf.image.adjust_brightness( examples[3], delta=0.3)
ax.imshow(img_adj_brightness)
## Column 5: cropping from image center
ax = fig.add_subplot(2,5,5)
ax.imshow(examples[4])
ax = fig.add_subplot(2,5,10)
ax.set_title('Central_crop\nand resize', size=15)
img_center_crop =tf.image.central_crop( examples[4], 0.7)
img_resized = tf.image.resize( img_center_crop, size=(218, 178) )
ax.imshow( img_resized.numpy().astype('uint8') )
plt.show()The following figure shows the results:
In the previous figure, the original images are shown in the first row and their transformed version in the second row. Note that for the first transformation (leftmost column), the bounding box is specified by four numbers: the coordinate of the upper-left corner of the bounding box (here x=20, y=50), and the width and height of the box (width=128, height=128). Also note that the origin (the coordinates at the location denoted as (0, 0)) for images loaded by TensorFlow (as well as other
packages such as imageio) is the upper-left corner of the image.
The transformations in the previous code block are deterministic. However, all such transformations can also be randomized, which is recommended for data augmentation during model training. For example, a random bounding box (where the coordinates of the upper-left corner are selected randomly) can be cropped from an image, an image can be randomly flipped along either the horizontal or vertical axes with a probability of 0.5, or the contrast of an image can be changed randomly, where the contrast_factor is selected at random, but with uniform distribution, from a range of values. In addition, we can create a pipeline of these transformations.
For example, we can first randomly crop an image, then flip it randomly, and finally, resize it to the desired size. The code is as follows (since we have random elements, we set the random seed for reproducibility):
tf.random.set_seed(1)
fig = plt.figure( figsize=(14,12) )
for i, example in enumerate( celeba_train.take(3) ):
image = example['image']
ax = fig.add_subplot(3,4, i*4+1)
ax.imshow( image )
if i ==0 :
ax.set_title('Orig.', size=15)
ax = fig.add_subplot(3,4, i*4+2)
img_crop = tf.image.random_crop( image, size=(178, 178,3) )
ax.imshow( img_crop )
if i==0:
ax.set_title('Step 1: Random crop', size=15)
ax = fig.add_subplot(3,4, i*4+3)
img_flip = tf.image.random_flip_left_right(img_crop)
ax.imshow( tf.cast(img_flip, tf.uint8) )###
if i==0:
ax.set_title('Step 2: Random flip', size=15)
ax = fig.add_subplot(3,4, i*4+4)
img_resized = tf.image.resize(img_flip, size=(128, 128))
ax.imshow( tf.cast(img_resized, tf.uint8) )
if i==0:
ax.set_title('Step 3: Resize', size=15)
plt.show()The following figure shows random transformations on three example images:
Note that each time we iterate through these three examples, we get slightly different images due to random transformations.
For convenience, we can define a wrapper function to use this pipeline for data augmentation during model training. In the following code, we will define the function preprocess(), which will receive a dictionary containing the keys 'image'
and 'attributes'. The function will return a tuple containing the transformed image and the label extracted from the dictionary of attributes.
We will only apply data augmentation to the training examples, however, and not to the validation or test images. The code is as follows:
def preprocess(example, size=(64, 64), mode='train'):
image = example['image']
label = example['attributes']['Male']
if mode == 'train':
image_cropped = tf.image.random_crop( image, size=(178,178,3) )
image_resized = tf.image.resize( image_cropped, size=size )
image_flip = tf.image.random_flip_left_right( image_resized )
return image_flip/255.0, tf.cast(label, tf.int32)
else: # use center- instead of random-crops for non-train data
image_cropped = tf.image.crop_to_bounding_box( image, 20, 0, 178, 178 )
image_resized = tf.image.resize(image_cropped, size=size )
return image_resized/255.0, tf.cast(label, tf.int32)
### testing:
# item = next(iter(celeba_train))
# preprocess(item, mode='train') Now, to see data augmentation in action, let's create a small subset of the training dataset, apply this function to it, and iterate over the dataset five times:
tf.random.set_seed(1)
ds = celeba_train.shuffle( 1000, reshuffle_each_iteration=False )
ds = ds.take(2).repeat(5) # [2,2,2,2,2]
ds = ds.map( lambda x: preprocess(x, size=(178,178), mode='train' ) )
fig = plt.figure( figsize=(15,6) )
for j,example in enumerate(ds):
ax = fig.add_subplot( 2,5, j//2 + (j%2)*5+1 )########
ax.set_xticks([])
ax.set_yticks([])
ax.imshow(example[0])This figure shows the five resulting transformations for data augmentation on two example images:
Next, we will apply this preprocessing function to our training and validation datasets. We will use an image size of (64, 64). Furthermore, we will specify mode='train' when working with training data and use mode='eval' for the validation data so that the random elements of the data augmentation pipeline will be applied only to the training data:
import numpy as np
BATCH_SIZE = 32
BUFFER_SIZE = 1000
IMAGE_SIZE = (64,64)
steps_per_epoch = np.ceil( 16000/BATCH_SIZE )
print(steps_per_epoch)
ds_train = celeba_train.map(
lambda x: preprocess(x, size=(IMAGE_SIZE), mode='train')
)
ds_train = ds_train.shuffle( buffer_size=BUFFER_SIZE ).repeat()
ds_train = ds_train.batch( BATCH_SIZE )
ds_valid = celeba_valid.map( lambda x: preprocess(x, size=IMAGE_SIZE, mode='eval') )
ds_valid = ds_valid.batch( BATCH_SIZE ) ![]()
Training a CNN gender classifier
By now, building a model with TensorFlow's Keras API and training it should be straightforward. The design of our CNN is as follows: the CNN model receives input images of size 64 × 64 × 3 (the images have three color channels, using 'channels_last').
The input data goes through four convolutional layers to make 32, 64, 128, and 256 feature maps using filters with kernel a size of 3 × 3 . The first three convolution layers are followed by max-pooling, ![]() . Two dropout layers are also included for regularization:
. Two dropout layers are also included for regularization:
model = tf.keras.Sequential([
tf.keras.layers.Conv2D( 32, (3,3), padding='same', activation='relu'), #if n=64 then output_shape=64, (64+2p-3)/1 +1 = 64 ==>p=1
tf.keras.layers.MaxPooling2D( (2,2) ), #strides: If None, it will default to `pool_size`.# (64+2*0-2)/2+1=32
tf.keras.layers.Dropout( rate=0.5 ),
tf.keras.layers.Conv2D( 64, (3,3), padding='same', activation='relu'), # output_shape=32
tf.keras.layers.MaxPooling2D( (2,2) ), # outputshape=16
tf.keras.layers.Dropout( rate=0.5 ),
tf.keras.layers.Conv2D( 128, (3,3), padding='same', activation='relu'),# outputshape=16
tf.keras.layers.MaxPooling2D( (2,2) ), # outputshape=8
tf.keras.layers.Conv2D( 256, (3,3), padding='same', activation='relu'),# outputshape=8
])Let's see the shape of the output feature maps after applying these layers:
model.compute_output_shape( input_shape=(None, 64,64,3) )![]()
There are 256 feature maps (or channels) of size 8 × 8 . Now, we can add a fully connected layer to get to the output layer with a single unit. If we reshape (flatten) the feature maps, the number of input units to this fully connected layer will be 8 × 8 × 256 = 16,384 . Alternatively, let's consider a new layer, called global averagepooling, which computes the average of each feature map separately, thereby reducing the hidden units to 256. We can then add a fully connected layer. Although we have not discussed global average-pooling explicitly明确地, it is conceptually very similar to other pooling layers. Global average-pooling can be viewed, in fact, as a special case of average-pooling when the pooling size is equal to the size of the input feature maps.
To understand this, consider the following figure showing an example of input feature maps of shape [batchsize × 64 × 64 × 8]. The channels are numbered = 0, 1, … , 7 . The global average-pooling operation calculates the average of each channel so that the output will have shape [batchsize × 8]. (Note: GlobalAveragePooling2D in the Keras API will automatically squeeze压缩 the output. Without squeezing the output, the shape would be [batchsize × 1 × 1 × 8]( ==>[batchsize × 8] ) , as the global average-pooling would reduce the spatial dimension of 64 × 64 to a 1 × 1) :
Therefore, given that, in our case, the shape of the feature maps prior to this layer is [batchsize × 8 × 8 × 256], we expect to get 256 units as output, that is, the shape of the output will be [batchsize × 256]. Let's add this layer and recompute the output shape to verify that this is true:
model.add( tf.keras.layers.GlobalAveragePooling2D() )
model.compute_output_shape( input_shape=(None, 64, 64, 3) )![]()
Finally, we can add a fully connected (dense) layer to get a single output unit. In this case, we can specify the activation function to be 'sigmoid' or just use activation=None, so that the model will output the logits (instead of classmembership
probabilities), which is preferred for model training in TensorFlow and Keras due to numerical stability, as discussed earlier:
model.add( tf.keras.layers.Dense(1, activation=None) )
tf.random.set_seed(1)
model.build( input_shape=(None, 64,64,3) )
model.summary() The next step is to compile the model and, at this time, we have to decide what loss function to use. We have a binary classification with a single output unit, so that means we should use BinaryCrossentropy. In addition, since our last layer does not apply sigmoid activation (we used activation=None), the outputs of our model are logits, not probabilities. Therefore, we will also specify from_logits=True in BinaryCrossentropy so that the loss function applies the sigmoid function
internally, which is, due to the underlying code, more efficient than doing it manually. The code for compiling and training the model is as follows:
from google.colab import drive
drive.mount('/content/gdrive') ![]()
tf.random.set_seed(1)
np.random.seed(1)
model.compile( optimizer=tf.keras.optimizers.Adam(),
loss=tf.keras.losses.BinaryCrossentropy(from_logits=True), #from_logits: due to numerical stability
metrics=['accuracy']
)
# https://www.tensorflow.org/api_docs/python/tf/keras/callbacks/ModelCheckpoint
# mode = "auto"
# monitor='val_loss' #default
filepath="/content/gdrive/My Drive/Colab Notebooks/CelebA_CNN_model.{epoch:02d}.hdf5"
checkpoint_cb = tf.keras.callbacks.ModelCheckpoint(filepath,
save_freq="epoch",
)
from tensorflow.keras.models import load_model
callbacks_list = [checkpoint_cb]
history = model.fit( ds_train,
validation_data=ds_valid,
epochs=20,
steps_per_epoch=steps_per_epoch, #steps_per_epoch = np.ceil( 16000/BATCH_SIZE )
callbacks=[callbacks_list]##########
)Let's now visualize the learning curve and compare the training and validation loss and accuracies after each epoch:
hist = history.history
x_arr = np.arange( len(hist['loss']) )+1
fig = plt.figure( figsize=(12,4) )
ax = fig.add_subplot(1,2,1)
ax.plot( x_arr, hist['loss'], '-o', label='Train loss')
ax.plot( x_arr, hist['val_loss'], '--<', label='Validation loss')
ax.legend( fontsize=15 )
ax.set_xlabel('Epoch', size=15)
ax.set_ylabel('Loss', size=15)
ax = fig.add_subplot(1,2,2)
ax.plot( x_arr, hist['accuracy'], '-o', label='Train acc.')
ax.plot( x_arr, hist['val_accuracy'], '--<', label='Validation acc.')
ax.legend( fontsize=15 )
ax.set_xlabel('Epoch', size=15)
ax.set_ylabel('Accuracy', size=15)
plt.show()The following figure shows the training and validation losses and accuracies:
As you can see from the learning curves, the losses for the training and validation have not converged to a plateau高原 region. Based on this result, we could have continued training for a few more epochs. Using the fit() method, we can continue training for an additional 10 epochs as follows:
import pickle
with open('/content/gdrive/My Drive/Colab Notebooks/trainHistory20', 'wb') as histFile:
pickle.dump( hist, histFile)
# with open('/content/gdrive/My Drive/Colab Notebooks/trainHistory20', 'rb') as histFile:
# hist20=pickle.load(histFile)
# hist20ds_test = celeba_test.map( lambda x: preprocess(x, size=IMAGE_SIZE, mode = 'eval') ).batch(32)
results = model.evaluate(ds_test) # verbose=0 # [loss, accuracy]
print( 'Test Acc: {:.2f}%'.format(results[1]*100) ) # results:[0.13623908162117004, 0.9386835098266602]model.save("/content/gdrive/My Drive/Colab Notebooks/CelebA_CNN_model.h5")
#model = tf.keras.models.load_model("/content/gdrive/My Drive/Colab Notebooks/CelebA_CNN_model.h5") ![]()
history = model.fit(ds_train,
validation_data=ds_valid,
epochs=30, initial_epoch=20,#continue training for an additional 10 epochs
steps_per_epoch=steps_per_epoch)hist2 = history.history
x_arr = np.arange( len( hist['loss']+hist2['loss'] ) )+1
fig = plt.figure( figsize=(12,4) )
ax = fig.add_subplot(1,2,1)
ax.plot( x_arr, hist['loss']+hist2['loss'], '-o', label='Train loss')
ax.plot( x_arr, hist['val_loss']+hist2['val_loss'], '--<', label='Validation loss')
ax.legend( fontsize=15 )
ax.set_xlabel('Epoch', size=15)
ax.set_ylabel('Loss', size=15)
ax = fig.add_subplot(1,2,2)
ax.plot( x_arr, hist['accuracy']+ hist2['accuracy'], '-o', label='Train acc.')
ax.plot( x_arr, hist['val_accuracy']+hist2['val_accuracy'], '--<', label='Validation acc.')
ax.legend( fontsize=15 )
ax.set_xlabel('Epoch', size=15)
ax.set_ylabel('Accuracy', size=15)
plt.show()import pickle
with open('/content/gdrive/My Drive/Colab Notebooks/trainHistory30', 'wb') as histFile:
pickle.dump( hist2, histFile)
# with open('/content/gdrive/My Drive/Colab Notebooks/trainHistory30', 'rb') as histFile:
# hist2=pickle.load(histFile)
# hist2 ds_test = celeba_test.map( lambda x: preprocess(x, size=IMAGE_SIZE, mode = 'eval') ).batch(32)
results = model.evaluate(ds_test) # verbose=0
print( 'Test Acc: {:.2f}%'.format(results[1]*100) )ds = ds_test.unbatch().take(10)
pred_logits = model.predict( ds.batch(10) )
probas = tf.sigmoid( pred_logits )
probas = probas.numpy().flatten()*100
fig = plt.figure( figsize=(15,7) )
for j, example in enumerate(ds):
ax = fig.add_subplot(2,5, j+1)
ax.set_xticks([])
ax.set_yticks([])
ax.imshow(example[0])
if example[1].numpy()==1: # example[1] : label
label='Male'
else:
label='Female'
ax.text( 0.5, -0.10,
'GT: {:s}\nPr(Male)={:.0f}%'.format(label, probas[j]),
size=16,
horizontalalignment='center',
verticalalignment='center',
transform=ax.transAxes )
plt.tight_layout()
plt.show() The probabilities of class 1 (that is, male according to CelebA) are provided below each image. As you can see, our trained model made only one error[0,2] on this set of 10 test examples.
The probabilities of class 1 (that is, male according to CelebA) are provided below each image. As you can see, our trained model made only one error[0,2] on this set of 10 test examples.
model = tf.keras.models.load_model("/content/gdrive/My Drive/Colab Notebooks/CelebA_CNN_model.h5")
history = model.fit(ds_train,
validation_data=ds_valid,
epochs=28, initial_epoch=20,######
steps_per_epoch=steps_per_epoch) ![]()
hist2 = history.history
x_arr = np.arange( len( hist['loss']+hist2['loss'] ) )+1
fig = plt.figure( figsize=(12,4) )
ax = fig.add_subplot(1,2,1)
ax.plot( x_arr, hist['loss']+hist2['loss'], '-o', label='Train loss')
ax.plot( x_arr, hist['val_loss']+hist2['val_loss'], '--<', label='Validation loss')
ax.legend( fontsize=15 )
ax.set_xlabel('Epoch', size=15)
ax.set_ylabel('Loss', size=15)
ax = fig.add_subplot(1,2,2)
ax.plot( x_arr, hist['accuracy']+ hist2['accuracy'], '-o', label='Train acc.')
ax.plot( x_arr, hist['val_accuracy']+hist2['val_accuracy'], '--<', label='Validation acc.')
ax.legend( fontsize=15 )
ax.set_xlabel('Epoch', size=15)
ax.set_ylabel('Accuracy', size=15)
plt.show()Once we are happy with the learning curves, we can evaluate the model on the holdout test dataset:
ds_test = celeba_test.map( lambda x: preprocess(x, size=IMAGE_SIZE, mode = 'eval') ).batch(32)
results = model.evaluate(ds_test) # verbose=0
print( 'Test Acc: {:.2f}%'.format(results[1]*100) )model.save('/content/gdrive/My Drive/Colab Notebooks/celeba-cnn-final28.h5')
import pickle
with open('/content/gdrive/My Drive/Colab Notebooks/trainHistoryFinal28', 'wb') as histFile:
pickle.dump( hist2, histFile)
# with open('/content/gdrive/My Drive/Colab Notebooks/trainHistoryFinal28', 'rb') as histFile:
# hist2=pickle.load(histFile)
# hist2
Finally, we already know how to get the prediction results on some test examples using model.predict(). However, remember that the model outputs the logits( since we used activation=None), not probabilities. If we are interested in the class-membership probabilities for this binary problem with a single output unit, we can use the tf.sigmoid 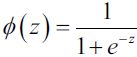 function to compute the probability of class 1. (In the case of a multiclass problem, we would use tf.math.softmax
function to compute the probability of class 1. (In the case of a multiclass problem, we would use tf.math.softmax![]() .)
.)
In the following code, we will take a small subset of 10 examples from our pre-processed test dataset (ds_test) and run model.predict() to get the logits. Then, we will compute the probabilities of each example being from class 1 (which corresponds to male based on the labels provided in CelebA) and visualize the examples along with their ground truth label and the predicted probabilities. Notice that we first apply unbatch() to the ds_test dataset before taking 10
examples; otherwise, the take() method would return 10 batches of size 32, instead of 10 individual examples:
In the following figure, you can see 10 example images along with their ground truth labels and the probabilities that they belong to class 1, male:
ds = ds_test.unbatch().take(10)
pred_logits = model.predict( ds.batch(10) )
probas = tf.sigmoid( pred_logits )
probas = probas.numpy().flatten()*100
fig = plt.figure( figsize=(15,7) )
for j, example in enumerate(ds):
ax = fig.add_subplot(2,5, j+1)
ax.set_xticks([])
ax.set_yticks([])
ax.imshow(example[0])
if example[1].numpy()==1: # example[1] : label
label='Male'
else:
label='Female'
ax.text( 0.5, -0.10,
'GT: {:s}\nPr(Male)={:.0f}%'.format(label, probas[j]),
size=16,
horizontalalignment='center',
verticalalignment='center',
transform=ax.transAxes )
plt.tight_layout()
plt.show() As an optional exercise, you are encouraged to try using the entire training dataset instead of the small subset we created. Furthermore, you can change or modify the CNN architecture. For example, you can change the dropout probabilities and the
number of filters in the different convolutional layers. Also, you could replace the global average-pooling with a dense layer. If you are using the entire training dataset with the CNN architecture we trained in this chapter, you should be able to achieve about 97-99% accuracy.
Summary
In this chapter, we learned about CNNs and their main components. We started with the convolution operation and looked at 1D and 2D implementations. Then, we covered another type of layer that is found in several common CNN architectures:
the subsampling or so-called pooling layers. We primarily focused on the two most common forms of pooling: max-pooling and average-pooling.
Next, putting all these individual concepts together, we implemented deep CNNs using the TensorFlow Keras API. The first network we implemented was applied to the already familiar MNIST handwritten digit recognition problem.
Then, we implemented a second CNN on a more complex dataset consisting of face images and trained the CNN for gender classification. Along the way, you also learned about data augmentation and different transformations that we can apply to
face images using the TensorFlow Dataset class.
In the next chapter, we will move on to recurrent neural networks (RNNs). RNNs are used for learning the structure of sequence data, and they have some fascinating applications, including language translation and image captioning.
Appendix:
The effect of initial shuffling
import tensorflow as tf
import tensorflow_datasets as tfds
import numpy as np
import pandas as pd
## MNIST dataset
#datasets = tfds.load(name='mnist')
mnist_bldr = tfds.builder('mnist') ##################
mnist_bldr.download_and_prepare()
datasets = mnist_bldr.as_dataset(shuffle_files=False)##################
mnist_train_orig, mnist_test_orig = datasets['train'], datasets['test']
mnist_train = mnist_train_orig.map(
lambda item: (tf.cast(item['image'], tf.float32)/255.0,
tf.cast(item['label'], tf.int32)))
mnist_test = mnist_test_orig.map(
lambda item: (tf.cast(item['image'], tf.float32)/255.0,
tf.cast(item['label'], tf.int32)))
tf.random.set_seed(1)
mnist_train = mnist_train.shuffle(buffer_size=10000,
reshuffle_each_iteration=False)
mnist_valid = mnist_train.take(100)#.batch(BATCH_SIZE)
mnist_train = mnist_train.skip(100)#.batch(BATCH_SIZE) Notice that count-of-labels in mnist_valid did stay the same when the dataset is loaded with using Builder and specifying mnist_bldr.as_dataset(shuffle_files=False)
from collections import Counter
def count_labels(ds):
counter = Counter()
for example in ds:
counter.update([example[1].numpy()])
return counter
print('Count of labels:', count_labels(mnist_valid))
print('Count of labels:', count_labels(mnist_valid))![]()
import tensorflow_datasets as tfds
import numpy as np
import pandas as pd
## MNIST dataset
datasets = tfds.load(name='mnist')
#mnist_bldr = tfds.builder('mnist')
#mnist_bldr.download_and_prepare()
#datasets = mnist_bldr.as_dataset(shuffle_files=False)
mnist_train_orig, mnist_test_orig = datasets['train'], datasets['test']
mnist_train = mnist_train_orig.map(
lambda item: (tf.cast(item['image'], tf.float32)/255.0,
tf.cast(item['label'], tf.int32)))
mnist_test = mnist_test_orig.map(
lambda item: (tf.cast(item['image'], tf.float32)/255.0,
tf.cast(item['label'], tf.int32)))
tf.random.set_seed(1)
mnist_train = mnist_train.shuffle(buffer_size=10000,
reshuffle_each_iteration=False)
mnist_valid = mnist_train.take(100)#.batch(BATCH_SIZE)
mnist_train = mnist_train.skip(100)#.batch(BATCH_SIZE)
from collections import Counter
def count_labels(ds):
counter = Counter()
for example in ds:
counter.update([example[1].numpy()])
return counter
print('Count of labels:', count_labels(mnist_valid))
print('Count of labels:', count_labels(mnist_valid))