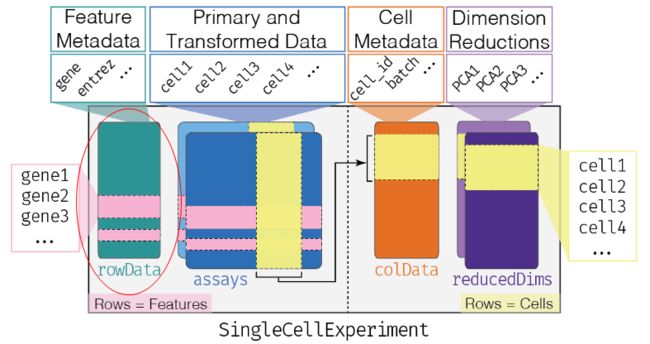
SingleCellExperiment是通过SingleCellExperiment包创建的单细胞数据分析对象,已有几十个单细胞R包支持。其衍生自SummarizedExperiment,之前在GEO数据挖掘学习时,了解过相关知识,主要是assay与pData两个函数的使用。
- 参考文章:https://osca.bioconductor.org/data-infrastructure.html
SingleCellExperiment
0、从创建一个SingleCellExperiment对象开始
library(SingleCellExperiment)
nm_count <- read.csv("GSE81861_CRC_NM_epithelial_cells_COUNT.csv")
row.names(nm_count) <- nm_count[,1]
nm_count <- nm_count[,-1]
nm_count <- as.matrix(nm_count)
rownames(nm_count) <- sapply(strsplit(rownames(nm_count),"_"),"[",2)
colnames(nm_count) <- sapply(strsplit(colnames(nm_count),"_"),"[",1)
nm_count[1:4,1:4]
dim(nm_count)
group.dat <- data.frame(group=rep("nomal",ncol(nm_count))) #不需要修改列名
sce <- SingleCellExperiment(assays = list(counts = nm_count),
colData = group.dat)
sce #查看对象的概况
1、assay:核心部分(盖了几层楼)
-
assays中最基本的是列为sample,行为feature(一般是gene)的表达矩阵。
如上代码,创建了名为counts的表达矩阵 - 后续可以根据需要,增添标准化、归一化等行列名不变的矩阵,相当于在count矩阵基础上“新盖的几层楼”。
1.1 查看矩阵
#根据矩阵名(楼层名)选择查看
assay(sce, "counts")[1:4,1:4]
#或者如下,但我觉得初学者使用第一种方法更能适应对象的结构
#counts(sce)[1:4,1:4]
assays(sce) #查看所有的层楼名,目前只有一个
1.2 新添矩阵
一般创建对象时都是引入counts矩阵。根据此基础矩阵可新建相关标准化等矩阵。
- 手动计算添加
counts <- assay(sce, "counts")
libsizes <- colSums(counts)
size.factors <- libsizes/mean(libsizes)
assay(sce, "logcounts") <- log2(t(t(counts)/size.factors) + 1)
assay(sce, "logcounts")[1:4,1:4]
assays(sce)
- 利用相关R包可自动添加
#注意下面自动添加的log标准化矩阵与上面的logcounts一致,可不运行
sce <- scater::logNormCounts(sce)
sce
assays(sce) #注意是assays,不是assay
assay(sce, "logcounts")[1:4,1:4]
关于assays的命名,我们可以随意命名。为了能够命名有意义并且配合R包之间的接口,官方给了如下的命名建议。
counts: Raw count data, e.g., number of reads or transcripts for a particular gene.
normcounts: Normalized values on the same scale as the original counts. For example, counts divided by cell-specific size factors that are centred at unity.
logcounts: Log-transformed counts or count-like values. In most cases, this will be defined as log-transformed normcounts, e.g., using log base 2 and a pseudo-count of 1.
cpm: Counts-per-million. This is the read count for each gene in each cell, divided by the library size of each cell in millions.
tpm: Transcripts-per-million. This is the number of transcripts for each gene in each cell, divided by the total number of transcripts in that cell (in millions).
2、colData:sample/cell annotation
- 本质上为行名为sample的data.frame
如上,创建对象时,引入了sample的group分组信息
colData(sce)
table(sce$group)
2.1 新增colData信息
-
$符可用于查看colData,也能用于新增。(这一点等同于dataframe)
sce$anything <- runif(ncol(sce))
colnames(colData(sce))
- scater包自动添加
sce <- scater::addPerCellQC(sce)
colData(sce)[, 1:5]
2.2 筛选sample
- 因为对象中所有的数据都是联动的,因此根据sample筛选出的sce中不含有筛选之外的所有信息。
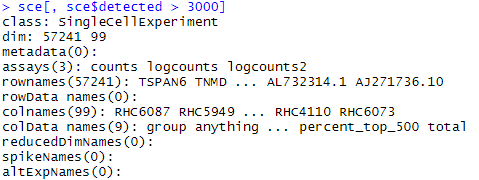
sce[, sce$detected > 3000]
sce #原来是160个sample
3、rowData:feature annotation
-参考colData,本质上为行名为feature的data.frame
- 可以在创建对象时添加(基因的不同ID),或者利用scater包添加注释。
sce <- scater::addPerFeatureQC(sce)
- 手动添加可用
cbind函数
rowData(x) <- cbind(rowData(x), feature)
#同理colData也类似,但使用$符也很方便
- 筛选feature
sce[rownames(nm_count)[c(1:10)], ]
sce[1:10, ]
Furthermore, there is a special
rowRangesslot to hold genomic coordinates in the form of aGRangesorGRangesList. This stores describes the chromosome, start, and end coordinates of the features (genes, genomic regions) in a manner that is easy to query and manipulate via the GenomicRanges framework.
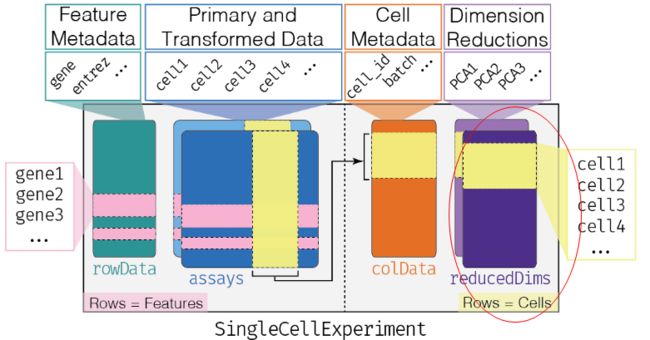
4、reducedDims
- 上面三点基本源于SummarizedExperiment对象,但reducedDims是sce对象独有的一点,一般储存PCA,tSNE,uMAP等细胞降维聚类信息;本质是行名为sample的一组data.frame
sce <- scater::logNormCounts(sce)
sce <- scater::runPCA(sce)
reducedDim(sce, "PCA")
reducedDim(sce, "PCA")[1:4,1:4]
# PC1 PC2 PC3 PC4
#RHC4104 46.66 -46.44 -9.488 1.79
#RHC6087 -54.38 -6.15 -0.155 13.16
#RHL2880 -7.89 -38.89 -1.319 -2.90
#RHC5949 -53.27 -20.88 -12.089 -7.73
sce <- scater::runTSNE(sce, perplexity = 0.1)
reducedDim(sce, "TSNE") #可用于绘图的二维坐标信息
reducedDims(sce) #类比assays
u <- uwot::umap(t(logcounts(sce)), n_neighbors = 2)
reducedDim(sce, "UMAP_uwot") <- u
reducedDims(sce) # Now stored in the object.
reducedDim(sce, "UMAP_uwot") #可用于绘图的二维坐标信息
5、other section
Alternative Experiments
- some data that have a distinct set of features but the same set of samples/cells.
- 即列名为sample,行名与assay不同类的表达矩阵,如spike-in transcripts、antibody-derived tags等
#官方示例代码,供以后参考
spike_counts <- cbind(cell_1 = rpois(5, 10),
cell_2 = rpois(5, 10),
cell_3 = rpois(5, 30))
rownames(spike_counts) <- paste0("spike_", 1:5)
spike_se <- SummarizedExperiment(list(counts=spike_counts))
spike_se
altExp(sce, "spike") <- spike_se
#引入失败,因为sample数目与名称不对
altExps(sce)
metadata
- 本质是一个列表list,储存一些记录等。
metadata(sce) <- list(GSE = 'GSE81861')
my_genes <- c("gene_1", "gene_5")
metadata(sce)$favorite_genes = my_genes
metadata(sce)