新版TCGA数据库学习:提取新版TCGA表达矩阵(tpm/count/fpkm)
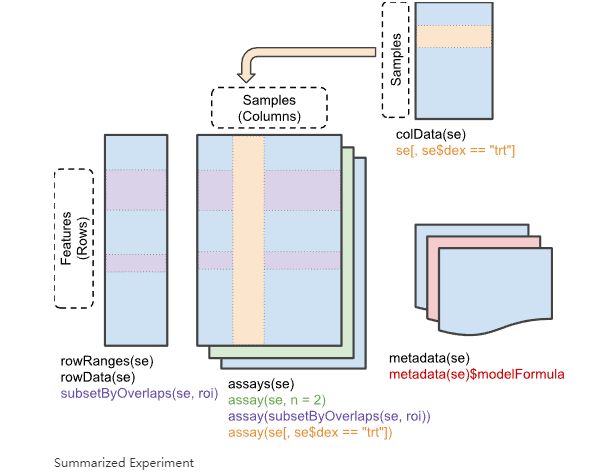
现在使用TCGAbiolinks下载转录组数据后,直接是一个SummarizedExperiment对象,这个对象非常重要且好用。因为里面直接包含了表达矩阵、样本信息、基因信息,可以非常方便的通过内置函数直接提取想要的数据,再也不用手扒了!!
是不是感觉和单细胞的SingCellExperiment对象非常像~
上次我们下载了常见的组学数据,今天学习下怎么提取数据,就以TCGA-READ的转录组数据为例。
分别提取mRNA和lncRNA的表达矩阵,还要添加gene symbol的那种!
加载数据和R包
加载之前下载好的数据。
rm(list = ls())
library(SummarizedExperiment)
## Loading required package: MatrixGenerics
## Loading required package: matrixStats
##
## Attaching package: 'MatrixGenerics'
## The following objects are masked from 'package:matrixStats':
##
## colAlls, colAnyNAs, colAnys, colAvgsPerRowSet, colCollapse,
## colCounts, colCummaxs, colCummins, colCumprods, colCumsums,
## colDiffs, colIQRDiffs, colIQRs, colLogSumExps, colMadDiffs,
## colMads, colMaxs, colMeans2, colMedians, colMins, colOrderStats,
## colProds, colQuantiles, colRanges, colRanks, colSdDiffs, colSds,
## colSums2, colTabulates, colVarDiffs, colVars, colWeightedMads,
## colWeightedMeans, colWeightedMedians, colWeightedSds,
## colWeightedVars, rowAlls, rowAnyNAs, rowAnys, rowAvgsPerColSet,
## rowCollapse, rowCounts, rowCummaxs, rowCummins, rowCumprods,
## rowCumsums, rowDiffs, rowIQRDiffs, rowIQRs, rowLogSumExps,
## rowMadDiffs, rowMads, rowMaxs, rowMeans2, rowMedians, rowMins,
## rowOrderStats, rowProds, rowQuantiles, rowRanges, rowRanks,
## rowSdDiffs, rowSds, rowSums2, rowTabulates, rowVarDiffs, rowVars,
## rowWeightedMads, rowWeightedMeans, rowWeightedMedians,
## rowWeightedSds, rowWeightedVars
## Loading required package: GenomicRanges
## Loading required package: stats4
## Loading required package: BiocGenerics
##
## Attaching package: 'BiocGenerics'
## The following objects are masked from 'package:stats':
##
## IQR, mad, sd, var, xtabs
## The following objects are masked from 'package:base':
##
## anyDuplicated, append, as.data.frame, basename, cbind, colnames,
## dirname, do.call, duplicated, eval, evalq, Filter, Find, get, grep,
## grepl, intersect, is.unsorted, lapply, Map, mapply, match, mget,
## order, paste, pmax, pmax.int, pmin, pmin.int, Position, rank,
## rbind, Reduce, rownames, sapply, setdiff, sort, table, tapply,
## union, unique, unsplit, which.max, which.min
## Loading required package: S4Vectors
##
## Attaching package: 'S4Vectors'
## The following objects are masked from 'package:base':
##
## expand.grid, I, unname
## Loading required package: IRanges
##
## Attaching package: 'IRanges'
## The following object is masked from 'package:grDevices':
##
## windows
## Loading required package: GenomeInfoDb
## Loading required package: Biobase
## Welcome to Bioconductor
##
## Vignettes contain introductory material; view with
## 'browseVignettes()'. To cite Bioconductor, see
## 'citation("Biobase")', and for packages 'citation("pkgname")'.
##
## Attaching package: 'Biobase'
## The following object is masked from 'package:MatrixGenerics':
##
## rowMedians
## The following objects are masked from 'package:matrixStats':
##
## anyMissing, rowMedians
load("TCGA-mRNA/TCGA-READ_mRNA.Rdata")
se <- data
这个se就是你的对象,含有coldata, rowdata, meta-data,以及最重要的assay,共有6个assay
探索SummarizedExperiment对象
se
## class: RangedSummarizedExperiment
## dim: 60660 177
## metadata(1): data_release
## assays(6): unstranded stranded_first ... fpkm_unstrand fpkm_uq_unstrand
## rownames(60660): ENSG00000000003.15 ENSG00000000005.6 ...
## ENSG00000288674.1 ENSG00000288675.1
## rowData names(10): source type ... hgnc_id havana_gene
## colnames(177): TCGA-AG-3580-01A-01R-0821-07
## TCGA-AF-2692-11A-01R-A32Z-07 ... TCGA-AG-3894-01A-01R-1119-07
## TCGA-AG-3574-01A-01R-0821-07
## colData names(107): barcode patient ... paper_vascular_invasion_present
## paper_vital_status
看看这个对象,它告诉你:
- 类型是:RangedSummarizedExperiment
- 维度:60660行,177列
- 6个assay以及它们的名字
- 表达矩阵的行名
- 行信息(也就是基因信息),比如gene id,gene name,gene type
- 表达矩阵的列名(也就是样本名)
- 列信息,也就是样本信息,比如生存时间、生存状态这些
太齐全了有没有!!
每个assay你可以理解为一个表达矩阵,我们需要的counts矩阵、TPM矩阵、FPKM矩阵就是其中一个~
# 查看每个assay的名字
names(assays(se))
## [1] "unstranded" "stranded_first" "stranded_second" "tpm_unstrand"
## [5] "fpkm_unstrand" "fpkm_uq_unstrand"
每个基因属于mRNA还是lncRNA存储在rowData中,这个rowData你可以理解为一个包含基因各种信息的数据框。
其中gene_type是基因类型,帮助我们区分到底是lncRNA还是mRNA,当然还包括很多其他类型。
# 提取rowData
rowdata <- rowData(se)
# 看看rowData包括哪些内容,可以看到里面有我们需要的gene_name和gene_type
names(rowdata)
## [1] "source" "type" "score" "phase" "gene_id"
## [6] "gene_type" "gene_name" "level" "hgnc_id" "havana_gene"
# gene_type是基因类型,看看有哪些
table(rowdata$gene_type)
##
## IG_C_gene IG_C_pseudogene
## 14 9
## IG_D_gene IG_J_gene
## 37 18
## IG_J_pseudogene IG_pseudogene
## 3 1
## IG_V_gene IG_V_pseudogene
## 145 187
## lncRNA miRNA
## 16901 1881
## misc_RNA Mt_rRNA
## 2212 2
## Mt_tRNA polymorphic_pseudogene
## 22 48
## processed_pseudogene protein_coding
## 10167 19962
## pseudogene ribozyme
## 18 8
## rRNA rRNA_pseudogene
## 47 497
## scaRNA scRNA
## 49 1
## snoRNA snRNA
## 943 1901
## sRNA TEC
## 5 1057
## TR_C_gene TR_D_gene
## 6 4
## TR_J_gene TR_J_pseudogene
## 79 4
## TR_V_gene TR_V_pseudogene
## 106 33
## transcribed_processed_pseudogene transcribed_unitary_pseudogene
## 500 138
## transcribed_unprocessed_pseudogene translated_processed_pseudogene
## 939 2
## translated_unprocessed_pseudogene unitary_pseudogene
## 1 98
## unprocessed_pseudogene vault_RNA
## 2614 1
gene_name就是gene_symbol,我们的id转换就用这一列信息。
# gene_name就是gene_symbol,就是我们需要的
head(rowdata$gene_name)
## [1] "TSPAN6" "TNMD" "DPM1" "SCYL3" "C1orf112" "FGR"
length(rowdata$gene_name)
## [1] 60660
还有一个重要的知识点是:SummarizedExperiment对象可以取子集,就像对数据框取子集那样,选择符合条件的行和列,并且子集也是SummarizedExperiment对象!
rowdata <- rowData(se)
# 分别提取mRNA和lncRNA的SummarizedExperiment对象
# 根据gene_type取子集,太简单了!
se_mrna <- se[rowdata$gene_type == "protein_coding",]
se_lnc <- se[rowdata$gene_type == "lncRNA"]
se_mrna #还是一个SummarizedExperiment对象,神奇!
## class: RangedSummarizedExperiment
## dim: 19962 177
## metadata(1): data_release
## assays(6): unstranded stranded_first ... fpkm_unstrand fpkm_uq_unstrand
## rownames(19962): ENSG00000000003.15 ENSG00000000005.6 ...
## ENSG00000288674.1 ENSG00000288675.1
## rowData names(10): source type ... hgnc_id havana_gene
## colnames(177): TCGA-AG-3580-01A-01R-0821-07
## TCGA-AF-2692-11A-01R-A32Z-07 ... TCGA-AG-3894-01A-01R-1119-07
## TCGA-AG-3574-01A-01R-0821-07
## colData names(107): barcode patient ... paper_vascular_invasion_present
## paper_vital_status
se_lnc
## class: RangedSummarizedExperiment
## dim: 16901 177
## metadata(1): data_release
## assays(6): unstranded stranded_first ... fpkm_unstrand fpkm_uq_unstrand
## rownames(16901): ENSG00000082929.8 ENSG00000083622.8 ...
## ENSG00000288667.1 ENSG00000288670.1
## rowData names(10): source type ... hgnc_id havana_gene
## colnames(177): TCGA-AG-3580-01A-01R-0821-07
## TCGA-AF-2692-11A-01R-A32Z-07 ... TCGA-AG-3894-01A-01R-1119-07
## TCGA-AG-3574-01A-01R-0821-07
## colData names(107): barcode patient ... paper_vascular_invasion_present
## paper_vital_status
提取表达矩阵
有了这些东西,就可以提取表达矩阵了,直接使用assay()搞定!
# mRNA的counts矩阵
expr_counts_mrna <- assay(se_mrna,"unstranded")
# mRNA的tpm矩阵
expr_tpm_mrna <- assay(se_mrna,"tpm_unstrand")
# mRNA的fpkm矩阵
expr_fpkm_mrna <- assay(se_mrna,"fpkm_unstrand")
# lncRNA的counts矩阵
expr_counts_lnc <- assay(se_lnc,"unstranded")
# lncRNA的tpm矩阵
expr_tpm_lnc <- assay(se_lnc,"tpm_unstrand")
# lncRNA的fpkm矩阵
expr_fpkm_lnc <- assay(se_lnc,"fpkm_unstrand")
简单!方便!快捷!
随便展示下:
expr_counts_mrna[1:10,1:2]
## TCGA-AG-3580-01A-01R-0821-07 TCGA-AF-2692-11A-01R-A32Z-07
## ENSG00000000003.15 3199 5839
## ENSG00000000005.6 6 91
## ENSG00000000419.13 828 1867
## ENSG00000000457.14 386 639
## ENSG00000000460.17 228 289
## ENSG00000000938.13 130 452
## ENSG00000000971.16 277 5170
## ENSG00000001036.14 1648 2946
## ENSG00000001084.13 823 2414
## ENSG00000001167.14 619 1487
是不是很简单?肯定比之前简单多了吧?
添加gene_symbol
添加gene_symbol也就非常简单了,只要提取gene_name这一列,然后和原来的表达矩阵合并即可!
# 先提取gene_name
symbol_mrna <- rowData(se_mrna)$gene_name
head(symbol_mrna)
## [1] "TSPAN6" "TNMD" "DPM1" "SCYL3" "C1orf112" "FGR"
symbol_lnc <- rowData(se_lnc)$gene_name
head(symbol_lnc)
## [1] "LINC01587" "AC000061.1" "AC016026.1" "IGF2-AS" "RRN3P2"
## [6] "AC087235.1"
和你喜欢的表达矩阵合并就行了:
expr_counts_mrna_symbol <- cbind(data.frame(symbol_mrna),
as.data.frame(expr_counts_mrna))
非常顺利~
但是呢,此时gene_symbol是有重复的,看上图中就有2个CD99,需要去重!
去重复也很简单,这里我们保留最大的那个。
suppressPackageStartupMessages(library(tidyverse))
expr_read <- expr_counts_mrna_symbol %>%
as_tibble() %>% # tibble不支持row name,我竟然才发现!
mutate(meanrow = rowMeans(.[,-1]), .before=2) %>%
arrange(desc(meanrow)) %>%
distinct(symbol_mrna,.keep_all=T) %>%
select(-meanrow) %>%
column_to_rownames(var = "symbol_mrna") %>%
as.data.frame()
不过还是要注意,gene_symbol是有重复的,需要去重复哦~
结果就变成大家最熟悉的表达矩阵了:
这样一个表达矩阵就搞定了!
本文由mdnice多平台发布