BraTS2021脑肿瘤分割实战
BraTS2021脑肿瘤分割实战
Brain Tumor Segmentation (BraTS) Challenge 2021 Homepage
github项目地址 brats-unet: UNet for brain tumor segmentation
BraTS是MICCAI所有比赛中历史最悠久的,到2021年已经连续举办了10年,参赛人数众多,是学习医学图像分割最前沿的平台之一。
1.数据准备
简介:
比赛方提供多机构、多参数多模态核磁共振成像(mpMRI)数据集,包括训练集(1251例)和验证集(219例)以及测试集(530例),
一共2000例患者的mpMRI扫描结果。其中训练集包含图像和分割标签,验证集和测试集没有分割标签,验证集被用于公共排行榜,测试集不公开,用作参赛者的最终排名评测。
四种模态数据:flair, t1ce, t1, t2,每个模态的数据大小都为 240 x 240 x 155,且共享分割标签。
分割标签:[0, 1, 2, 4]
- label0:背景(bachground)
- label1:坏疽(NT, necrotic tumor core)
- label2:浮肿区域(ED,peritumoral edema)
- label4:增强肿瘤区域(ET,enhancing tumor)
本次比赛包括两个任务:
- Task1:mpMRI扫描中分割内在异质性脑胶质母细胞瘤区域
- Task2:预测术前基线扫描中的MGMT启动子甲基化状态
本文从数据处理、评价指标、损失函数、模型训练四个方面介绍Task1的整体实现过程
数据集下载地址:
1.官网:BraTS 2021 Challenge 需要注册和申请(包括训练集和验证集)
2.Kaggle:BRaTS 2021 Task 1 Dataset 建议在kaggle上下载,数据集与官网一致(不包括验证集)
数据准备:
下载数据集,解压后如下图所示:
每个病例包含四种模态的MRI图像和分割标签,结构如下:
BraTS2021_00000
├── BraTS2021_00000_flair.nii.gz
├── BraTS2021_00000_seg.nii.gz
├── BraTS2021_00000_t1ce.nii.gz
├── BraTS2021_00000_t1.nii.gz
└── BraTS2021_00000_t2.nii.gz
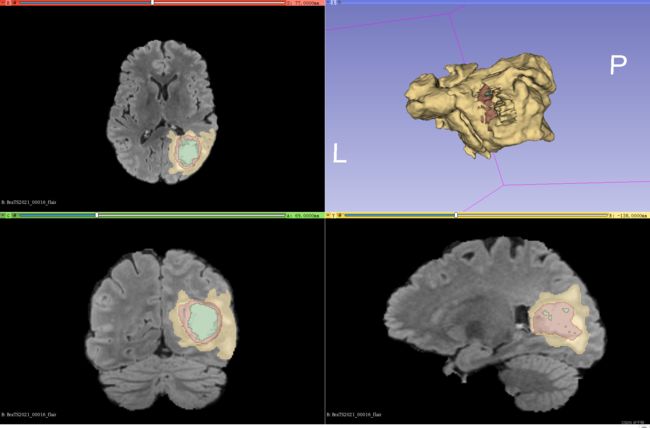
建议使用3D Slicer查看图像和标签,直观的了解一下自己要用的数据集。
2.数据预处理
每个病例的四种MRI图像大小为 240 x 240 x 155,且共享标签。
鉴于此,我将四种模态的图像合并为一个4D图像(H x W x D x C , C=4),并且和分割标签一起保存为一个.pkl文件,方便后续处理。
import pickle
import os
import numpy as np
import nibabel as nib
from tqdm import tqdm
# 四种模态的mri图像
modalities = ('flair', 't1ce', 't1', 't2')
# train
train_set = {
'root': '../data', # 训练集地址
'flist': 'train.txt', # 训练集列表
'has_label': True
}
def nib_load(file_name):
if not os.path.exists(file_name):
print('Invalid file name, can not find the file!')
proxy = nib.load(file_name) # 加载.nii.gz图像
data = proxy.get_data() # 获取图像数据
proxy.uncache()
return data
- 将数据保存为32位浮点数(np.float32),写入
.pkl文件 - 对每张图像的灰度进行标准化,但保持背景区域为0
def process_f32b0(path, has_label=True):
if has_label:
label = np.array(nib_load(path + 'seg.nii.gz'), dtype='uint8', order='C')
# 堆叠四种模态的图像,4 x (H,W,D) -> (H,W,D,4)
images = np.stack([np.array(nib_load(path + modal + '.nii.gz'), dtype='float32', order='C') for modal in modalities], -1) # [240,240,155]
# path是自定义的输出路径
path = '../BraTS2021/dataset/'+ path.split('/')[-1]
output = path + 'data_f32b0.pkl'
# 对最后一个通道求和,如果四个模态都为0,则标记为背景(False)
mask = images.sum(-1) > 0
for k in range(4):
x = images[..., k]
y = x[mask]
# 对背景外的区域进行归一化
x[mask] -= y.mean()
x[mask] /= y.std()
images[..., k] = x
with open(output, 'wb') as f:
if has_label:
pickle.dump((images, label), f) # 写入文件
else:
pickle.dump(images, f)
if not has_label:
return
def doit(dset):
root, has_label = dset['root'], dset['has_label']
file_list = os.path.join(root, dset['flist'])
subjects = open(file_list).read().splitlines()
names = ['BraTS2021_' + sub for sub in subjects]
paths = [os.path.join(root, name, name + '_') for name in names]
for path in tqdm(paths):
process_f32b0(path, has_label)
print('Finished')
if __name__ == '__main__':
doit(train_set)
将数据转换为.pkl文件读写速度快,缺点是占内存,注意预留足够的存储空间(大概181G,建议在服务器上处理)。若空间不足,可以将数据预处理和数据增强合并在一起进行在线处理。
处理后的数据,可以用下面的几行代码测试一下,记得修改为你自己的路径
import pickle
import numpy as np
def pkload(fname):
with open(fname, 'rb') as f:
return pickle.load(f)
path0 = '../BraTS2021/data/BraTS2021_00000_data_f32b0.pkl'
image,label = pkload(path0)
print('image shape:',image.shape,'\t','label shape',label.shape)
print('label set:',np.unique(label))
# image shape: (240, 240, 155, 4) label shape (240, 240, 155)
# label set: [0,1,2,4]
将数据集按照 8:1:1随机划分为训练集、验证集和测试集,将划分后的数据名保存为.txt文件
import os
from sklearn.model_selection import train_test_split
# 上一步处理后的数据集地址
data_path = "***/BraTS/dataset/data"
train_and_test_ids = os.listdir(data_path)
# random_state是随机种子数,建议自取
train_ids, val_test_ids = train_test_split(train_and_test_ids, test_size=0.2,random_state=21) # 8:2
val_ids, test_ids = train_test_split(val_test_ids, test_size=0.5,random_state=21) # 1:1
print("Using {} images for training, {} images for validation, {} images for testing.".format(len(train_ids),len(val_ids),len(test_ids)))
with open('***/BraTS/dataset/data/train.txt','w') as f:
f.write('\n'.join(train_ids))
with open('***/BraTS/dataset/valid.txt','w') as f:
f.write('\n'.join(val_ids))
with open('***/BraTS/dataset/test.txt','w') as f:
f.write('\n'.join(test_ids))
3.数据增强
下面是我写的Dataset类以及一些数据增强方法
整体架构
import os
import torch
from torch.utils.data import Dataset
import random
import numpy as np
from torchvision.transforms import transforms
import pickle
from scipy import ndimage
def pkload(fname):
with open(fname, 'rb') as f:
return pickle.load(f)
def transform(sample): # 训练集
trans = transforms.Compose([
Random_Crop(), # 裁剪
Random_rotate(), # 旋转
Random_Flip(), # 翻转
GaussianNoise(p=0.1), # 高斯噪声
ContrastAugmentationTransform(p_per_sample=0.15), # 对比度增强
BrightnessTransform(0,0.1,True,0.15,0.5), # 亮度变换
ToTensor() # 数据类型转换
])
return trans(sample)
def transform_valid(sample): # 验证集
trans = transforms.Compose([
Random_Crop(), # 裁剪
ToTensor() # 数据类型转换
])
return trans(sample)
class BraTS(Dataset):
def __init__(self,file_path,data_path="../BraTS2021/data", mode='train'):
with open(file_path, 'r') as f:
self.paths = [os.path.join(data_path, x.strip()) for x in f.readlines()]
self.mode = mode
def __getitem__(self, item):
path = self.paths[item]
if self.mode == 'train':
image, label = pkload(path)
# [h,w,s,c] -> [c,h,w,s]
image = image.transpose(3, 0, 1, 2)
sample = {'image': image, 'label': label}
sample = transform(sample)
return sample['image'], sample['label']
elif self.mode == 'valid':
image, label = pkload(path)
image = image.transpose(3, 0, 1, 2)
sample = {'image': image, 'label': label}
sample = transform_valid(sample)
return sample['image'], sample['label']
else:
image = pkload(path)
image = np.pad(image, ((0, 0), (0, 0), (0, 5), (0, 0)), mode='constant')
image = np.ascontiguousarray(image.transpose(3, 0, 1, 2))
image = torch.from_numpy(image).float()
return image
def __len__(self):
return len(self.paths)
def collate(self, batch):
return [torch.cat(v) for v in zip(*batch)]
if __name__ == '__main__':
data_path = "***/BraTS2021/data"
test_txt = "***/BraTS2021/test.txt"
test_set = BraTS(test_txt,data_path,'train')
# print(test_set.paths)
d1 = test_set[0]
image,label = d1
print(image.shape, label.shape, np.unique(label))
具体的数据增强方法我列在了下面,包括裁剪、旋转、翻转、高斯噪声、对比度变换和亮度增强的源码,部分代码借鉴了nnUNet的数据增强方法。
裁剪
原始图像尺寸为 240 x 240 x 155,但图像周围是有很多黑边的,我将图像裁剪为 160 x 160 x 128
我在D方向做的随机裁剪,实验发现随机裁剪比中心裁剪的训练结果要好
class Random_Crop(object):
"""
纵向随机裁剪
"""
def __call__(self, sample):
image = sample['image']
label = sample['label']
D = random.randint(0, 155 - 128)
# [4,240,240,155] -> [4,160,160,128]
image = image[:,40:200,40:200, D: D + 128]
label = label[40:200,40:200, D: D + 128]
return {'image': image, 'label': label}
旋转
任意角度的旋转可能会导致图像重采样,因为数据集比较充分,我只在{90,180,270}度做一个简单旋转,不涉及重采样。
class Random_rotate(object):
def __call__(self, sample):
image = sample['image']
label = sample['label']
angles = [90,180,270]
index = random.randint(0,2) # 0,1,2
# 在H,W所在的平面中随机旋转90,180,270度
image = ndimage.rotate(image, angles[index], axes=(1, 2), reshape=False)
label = ndimage.rotate(label, angles[index], axes=(0, 1), reshape=False)
return {'image': image, 'label': label}
翻转
class Random_Flip(object):
def __call__(self, sample):
image = sample['image']
label = sample['label']
if random.random() < 0.5:
image = np.flip(image, 1)
label = np.flip(label, 0)
if random.random() < 0.5:
image = np.flip(image, 2)
label = np.flip(label, 1)
if random.random() < 0.5:
image = np.flip(image, 3)
label = np.flip(label, 2)
return {'image': image, 'label': label}
高斯噪声
def augment_gaussian_noise(data_sample, noise_variance=(0, 0.1)):
if noise_variance[0] == noise_variance[1]:
variance = noise_variance[0]
else:
variance = random.uniform(noise_variance[0], noise_variance[1])
data_sample = data_sample + np.random.normal(0.0, variance, size=data_sample.shape)
return data_sample
class GaussianNoise(object):
"""
加性高斯噪声
noise_variance:高斯噪声的方差分布
"""
def __init__(self, noise_variance=(0, 0.1), p=0.5):
self.prob = p
self.noise_variance = noise_variance
def __call__(self, sample):
image = sample['image']
label = sample['label']
if np.random.uniform() < self.prob:
image = augment_gaussian_noise(image, self.noise_variance)
return {'image': image, 'label': label}
对比度变换
- contrast_range:对比度增强的范围
- preserve_range:是否保留数据的取值范围
- per_channel:是否对每个通道的图像分别进行对比度增强
def augment_contrast(data_sample, contrast_range=(0.75, 1.25), preserve_range=True, per_channel=True):
if not per_channel:
mn = data_sample.mean()
if preserve_range:
minm = data_sample.min()
maxm = data_sample.max()
if np.random.random() < 0.5 and contrast_range[0] < 1:
factor = np.random.uniform(contrast_range[0], 1)
else:
factor = np.random.uniform(max(contrast_range[0], 1), contrast_range[1])
data_sample = (data_sample - mn) * factor + mn
if preserve_range:
data_sample[data_sample < minm] = minm
data_sample[data_sample > maxm] = maxm
else:
for c in range(data_sample.shape[0]):
mn = data_sample[c].mean()
if preserve_range:
minm = data_sample[c].min()
maxm = data_sample[c].max()
if np.random.random() < 0.5 and contrast_range[0] < 1:
factor = np.random.uniform(contrast_range[0], 1)
else:
factor = np.random.uniform(max(contrast_range[0], 1), contrast_range[1])
data_sample[c] = (data_sample[c] - mn) * factor + mn
if preserve_range:
data_sample[c][data_sample[c] < minm] = minm
data_sample[c][data_sample[c] > maxm] = maxm
return data_sample
class ContrastAugmentationTransform(object):
def __init__(self, contrast_range=(0.75, 1.25), preserve_range=True, per_channel=True,p_per_sample=1.):
self.p_per_sample = p_per_sample
self.contrast_range = contrast_range
self.preserve_range = preserve_range
self.per_channel = per_channel
def __call__(self, sample):
image = sample['image']
label = sample['label']
for b in range(len(image)):
if np.random.uniform() < self.p_per_sample:
image[b] = augment_contrast(image[b], contrast_range=self.contrast_range,
preserve_range=self.preserve_range, per_channel=self.per_channel)
return {'image': image, 'label': label}
亮度变换
附加亮度从具有μ和σ的高斯分布中采样
def augment_brightness_additive(data_sample, mu:float, sigma:float , per_channel:bool=True, p_per_channel:float=1.):
if not per_channel:
rnd_nb = np.random.normal(mu, sigma)
for c in range(data_sample.shape[0]):
if np.random.uniform() <= p_per_channel:
data_sample[c] += rnd_nb
else:
for c in range(data_sample.shape[0]):
if np.random.uniform() <= p_per_channel:
rnd_nb = np.random.normal(mu, sigma)
data_sample[c] += rnd_nb
return data_sample
class BrightnessTransform(object):
def __init__(self, mu, sigma, per_channel=True, p_per_sample=1., p_per_channel=1.):
self.p_per_sample = p_per_sample
self.mu = mu
self.sigma = sigma
self.per_channel = per_channel
self.p_per_channel = p_per_channel
def __call__(self, sample):
data, label = sample['image'], sample['label']
for b in range(data.shape[0]):
if np.random.uniform() < self.p_per_sample:
data[b] = augment_brightness_additive(data[b], self.mu, self.sigma, self.per_channel,
p_per_channel=self.p_per_channel)
return {'image': data, 'label': label}
数据类型转换
将Numpy数组转为Tensor
class ToTensor(object):
"""Convert ndarrays in sample to Tensors."""
def __call__(self, sample):
image = sample['image']
label = sample['label']
image = np.ascontiguousarray(image)
label = np.ascontiguousarray(label)
image = torch.from_numpy(image).float()
label = torch.from_numpy(label).long()
return {'image': image, 'label': label}
相比其他医学影像数据集,BraTS2021是非常高质量的,对数据增强方法并不是很敏感。
4.评价损失
损失函数:
combination of dice and crossentropy loss
![]()
dice loss
![]()
- μ是网络的softmax输出
- v是分割标签的one-hot编码
其实就是将计算dice时的
torch.argmax替换为了torch.softmax
import torch.nn.functional as F
import torch.nn as nn
import torch
from einops import rearrange
class Loss(nn.Module):
def __init__(self, n_classes, weight=None, alpha=0.5):
"dice_loss_plus_cetr_weighted"
super(Loss, self).__init__()
self.n_classes = n_classes
self.weight = weight.cuda()
# self.weight = weight
self.alpha = alpha
def forward(self, input, target):
smooth = 0.01 # 防止分母为0
input1 = F.softmax(input, dim=1)
target1 = F.one_hot(target,self.n_classes)
input1 = rearrange(input1,'b n h w s -> b n (h w s)')
target1 = rearrange(target1,'b h w s n -> b n (h w s)')
input1 = input1[:, 1:, :]
target1 = target1[:, 1:, :].float()
# 以batch为单位计算loss和dice_loss,据说训练更稳定,和上面的公式有出入
# 注意,这里的dice不是真正的dice,叫做soft_dice更贴切
inter = torch.sum(input1 * target1)
union = torch.sum(input1) + torch.sum(target1) + smooth
dice = 2.0 * inter / union
loss = F.cross_entropy(input,target, weight=self.weight)
total_loss = (1 - self.alpha) * loss + (1 - dice) * self.alpha
return total_loss
if __name__ == '__main__':
torch.manual_seed(3)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
losser = Loss(n_classes=4, weight=torch.tensor([0.2, 0.3, 0.25, 0.25])).to(device)
x = torch.randn((2, 4, 16, 16, 16)).to(device)
y = torch.randint(0, 4, (2, 16, 16, 16)).to(device)
print(losser(x, y))
评价指标:
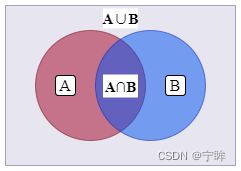
dice计算方法:
2 ( A ∩ B ) A ∪ B 2{(A \cap B)}\over{A \cup B} A∪B2(A∩B)
def Dice(output, target, eps=1e-3):
inter = torch.sum(output * target,dim=(1,2,-1)) + eps
union = torch.sum(output,dim=(1,2,-1)) + torch.sum(target,dim=(1,2,-1)) + eps * 2
x = 2 * inter / union
dice = torch.mean(x)
return dice
- output: (b, num_class, d, h, w) target: (b, d, h, w)
- dice1(ET):label4
- dice2(TC):label1 + label4
- dice3(WT): label1 + label2 + label4
- 注意,这里的label4已经被替换为3
def cal_dice(output, target):
output = torch.argmax(output,dim=1)
dice1 = Dice((output == 3).float(), (target == 3).float())
dice2 = Dice(((output == 1) | (output == 3)).float(), ((target == 1) | (target == 3)).float())
dice3 = Dice((output != 0).float(), (target != 0).float())
return dice1, dice2, dice3
5.模型训练
以UNet为例,我把完整代码放在了下面
module:
import torch
import torch.nn as nn
class InConv(nn.Module):
def __init__(self, in_ch, out_ch):
super(InConv, self).__init__()
self.conv = DoubleConv(in_ch, out_ch)
def forward(self, x):
x = self.conv(x)
return x
class Down(nn.Module):
def __init__(self, in_ch, out_ch):
super(Down, self).__init__()
self.mpconv = nn.Sequential(
nn.MaxPool3d(2, 2),
DoubleConv(in_ch, out_ch)
)
def forward(self, x):
x = self.mpconv(x)
return x
class OutConv(nn.Module):
def __init__(self, in_ch, out_ch):
super(OutConv, self).__init__()
self.conv = nn.Conv3d(in_ch, out_ch, 1)
# self.sigmoid = nn.Sigmoid()
def forward(self, x):
x = self.conv(x)
# x = self.sigmoid(x)
return x
class DoubleConv(nn.Module):
def __init__(self, in_ch, out_ch):
super(DoubleConv, self).__init__()
self.conv = nn.Sequential(
nn.Conv3d(in_ch, out_ch, kernel_size=3, stride=1, padding=1),
nn.BatchNorm3d(out_ch),
nn.ReLU(inplace=True),
nn.Conv3d(out_ch, out_ch, kernel_size=3, stride=1, padding=1),
nn.BatchNorm3d(out_ch),
nn.ReLU(inplace=True)
)
def forward(self, x):
x = self.conv(x)
return x
class Up(nn.Module):
def __init__(self, in_ch, skip_ch,out_ch):
super(Up, self).__init__()
self.up = nn.ConvTranspose3d(in_ch, in_ch, kernel_size=2, stride=2)
self.conv = DoubleConv(in_ch+skip_ch, out_ch)
def forward(self, x1, x2):
x1 = self.up(x1)
x = torch.cat([x2, x1], dim=1)
x = self.conv(x)
return x
model:
class UNet(nn.Module):
def __init__(self, in_channels, num_classes):
super(UNet, self).__init__()
features = [32,64,128,256]
self.inc = InConv(in_channels, features[0])
self.down1 = Down(features[0], features[1])
self.down2 = Down(features[1], features[2])
self.down3 = Down(features[2], features[3])
self.down4 = Down(features[3], features[3])
self.up1 = Up(features[3], features[3], features[2])
self.up2 = Up(features[2], features[2], features[1])
self.up3 = Up(features[1], features[1], features[0])
self.up4 = Up(features[0], features[0], features[0])
self.outc = OutConv(features[0], num_classes)
def forward(self, x):
x1 = self.inc(x)
x2 = self.down1(x1)
x3 = self.down2(x2)
x4 = self.down3(x3)
x5 = self.down4(x4)
x = self.up1(x5, x4)
x = self.up2(x, x3)
x = self.up3(x, x2)
x = self.up4(x, x1)
x = self.outc(x)
return x
if __name__ == '__main__':
x = torch.randn(1, 4, 160, 160, 128)
net = UNet(in_channels=4, num_classes=4)
y = net(x)
print("params: ", sum(p.numel() for p in net.parameters()))
print(y.shape)
Train:
下面是我写的训练函数,具体细节见代码注释
- 优化器:
optim.SGD(model.parameters(),momentum=0.9, lr=0, weight_decay=5e-4) - 学习率余弦衰减:最大学习率0.004,最小学习率0.002,预热10个epoch
- 优化策略可参考我的另一篇博客nnUnet代码解读–优化策略
import os
import argparse
from torch.utils.data import DataLoader
import torch
import torch.optim as optim
from tqdm import tqdm
from BraTS import BraTS
from Unet_models.Unet import UNet
from loss import Loss,cal_dice
from utils import cosine_scheduler
def train_loop(model,optimizer,scheduler,criterion,train_loader,device,epoch):
model.train()
running_loss = 0
dice1_train = 0
dice2_train = 0
dice3_train = 0
pbar = tqdm(train_loader)
for it,(images,masks) in enumerate(pbar):
# update learning rate according to the schedule
it = len(train_loader) * epoch + it
param_group = optimizer.param_groups[0]
param_group['lr'] = scheduler[it]
# print(scheduler[it])
# [b,4,160,160,128] , [b,160,160,128]
images, masks = images.to(device),masks.to(device)
# [b,4,160,160,128], 4分割
outputs = model(images)
loss = criterion(outputs, masks)
dice1, dice2, dice3 = cal_dice(outputs,masks)
pbar.desc = "loss: {:.3f} ".format(loss.item())
running_loss += loss.item()
dice1_train += dice1.item()
dice2_train += dice2.item()
dice3_train += dice3.item()
optimizer.zero_grad()
loss.backward()
optimizer.step()
loss = running_loss / len(train_loader)
dice1 = dice1_train / len(train_loader)
dice2 = dice2_train / len(train_loader)
dice3 = dice3_train / len(train_loader)
return {'loss':loss,'dice1':dice1,'dice2':dice2,'dice3':dice3}
def val_loop(model,criterion,val_loader,device):
model.eval()
running_loss = 0
dice1_val = 0
dice2_val = 0
dice3_val = 0
pbar = tqdm(val_loader)
with torch.no_grad():
for images, masks in pbar:
images, masks = images.to(device), masks.to(device)
outputs = model(images)
loss = criterion(outputs, masks)
dice1, dice2, dice3 = cal_dice(outputs, masks)
running_loss += loss.item()
dice1_val += dice1.item()
dice2_val += dice2.item()
dice3_val += dice3.item()
# pbar.desc = "loss:{:.3f} dice1:{:.3f} dice2:{:.3f} dice3:{:.3f} ".format(loss,dice1,dice2,dice3)
loss = running_loss / len(val_loader)
dice1 = dice1_val / len(val_loader)
dice2 = dice2_val / len(val_loader)
dice3 = dice3_val / len(val_loader)
return {'loss':loss,'dice1':dice1,'dice2':dice2,'dice3':dice3}
def train(model,optimizer,scheduler,criterion,train_loader,
val_loader,epochs,device,train_log,valid_loss_min=999.0):
for e in range(34,epochs):
# train for epoch
train_metrics = train_loop(model,optimizer,scheduler,criterion,train_loader,device,e)
# eval for epoch
val_metrics = val_loop(model,criterion,val_loader,device)
info1 = "Epoch:[{}/{}] train_loss: {:.3f} valid_loss: {:.3f} ".format(e+1,epochs,train_metrics["loss"],val_metrics["loss"])
info2 = "Train--ET: {:.3f} TC: {:.3f} WT: {:.3f} ".format(train_metrics['dice1'],train_metrics['dice2'],train_metrics['dice3'])
info3 = "Valid--ET: {:.3f} TC: {:.3f} WT: {:.3f} ".format(val_metrics['dice1'],val_metrics['dice2'],val_metrics['dice3'])
print(info1)
print(info2)
print(info3)
with open(train_log,'a') as f:
f.write(info1 + '\n' + info2 + ' ' + info3 + '\n')
if not os.path.exists(args.save_path):
os.makedirs(args.save_path)
save_file = {"model": model.state_dict(),
"optimizer": optimizer.state_dict()}
if val_metrics['loss'] < valid_loss_min:
valid_loss_min = val_metrics['loss']
torch.save(save_file, 'results/UNet.pth')
else:
torch.save(save_file,os.path.join(args.save_path,'checkpoint{}.pth'.format(e+1)))
print("Finished Training!")
def main(args):
torch.manual_seed(args.seed) # 为CPU设置种子用于生成随机数,以使得结果是确定的
torch.cuda.manual_seed_all(args.seed) # 为所有的GPU设置种子,以使得结果是确定的
torch.backends.cudnn.deterministic = True
torch.backends.cudnn.benchmark = True
os.environ['CUDA_VISIBLE_DEVICES'] = '0'
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
# data info
train_dataset = BraTS(args.train_txt,args.data_path, mode='train')
val_dataset = BraTS(args.valid_txt,args.data_path,mode='valid')
test_dataset = BraTS(args.test_txt,args.data_path,mode='valid')
train_loader = DataLoader(dataset=train_dataset, batch_size=args.batch_size, num_workers=12,
shuffle=True, pin_memory=True)
val_loader = DataLoader(dataset=val_dataset, batch_size=args.batch_size, num_workers=12, shuffle=False,
pin_memory=True)
test_loader = DataLoader(dataset=test_dataset, batch_size=args.batch_size, num_workers=12, shuffle=False,
pin_memory=True)
print("using {} device.".format(device))
print("using {} images for training, {} images for validation.".format(len(train_dataset), len(val_dataset)))
# img,label = train_dataset[0]
# 1-坏疽(NT,necrotic tumor core),2-浮肿区域(ED,peritumoral edema),4-增强肿瘤区域(ET,enhancing tumor)
# 评价指标:ET(label4),TC(label1+label4),WT(label1+label2+label4)
model = UNet(in_channels=4,num_classes=4,feature_scale=3).to(device)
criterion = Loss(n_classes=4, weight=torch.tensor([0.2, 0.3, 0.25, 0.25])).to(device)
optimizer = optim.SGD(model.parameters(),momentum=0.9, lr=0, weight_decay=5e-4)
scheduler = cosine_scheduler(base_value=args.lr,final_value=args.min_lr,epochs=args.epochs,
niter_per_ep=len(train_loader),warmup_epochs=args.warmup_epochs,start_warmup_value=5e-4)
# 加载训练模型
if os.path.exists(args.weights):
weight_dict = torch.load(args.weights, map_location=device)
model.load_state_dict(weight_dict['model'])
optimizer.load_state_dict(weight_dict['optimizer'])
print('Successfully loading checkpoint.')
train(model,optimizer,scheduler,criterion,train_loader,val_loader,args.epochs,device,train_log=args.train_log)
# metrics1 = val_loop(model, criterion, train_loader, device)
metrics2 = val_loop(model, criterion, val_loader, device)
metrics3 = val_loop(model, criterion, test_loader, device)
# 最后再评价一遍所有数据,注意,这里使用的是训练结束的模型参数
# 若想评价最好的训练结果,需要把上面的train函数注释掉,加载对应的模型参数
# print("Train -- loss: {:.3f} ET: {:.3f} TC: {:.3f} WT: {:.3f}".format(metrics1['loss'], metrics1['dice1'],metrics1['dice2'], metrics1['dice3']))
print("Valid -- loss: {:.3f} ET: {:.3f} TC: {:.3f} WT: {:.3f}".format(metrics2['loss'], metrics2['dice1'], metrics2['dice2'], metrics2['dice3']))
print("Test -- loss: {:.3f} ET: {:.3f} TC: {:.3f} WT: {:.3f}".format(metrics3['loss'], metrics3['dice1'], metrics3['dice2'], metrics3['dice3']))
if __name__ == '__main__':
parser = argparse.ArgumentParser()
parser.add_argument('--num_classes', type=int, default=4)
parser.add_argument('--seed', type=int, default=21)
parser.add_argument('--epochs', type=int, default=60)
parser.add_argument('--warmup_epochs', type=int, default=10)
parser.add_argument('--batch_size', type=int, default=5)
parser.add_argument('--lr', type=float, default=0.004)
parser.add_argument('--min_lr', type=float, default=0.002)
parser.add_argument('--data_path', type=str, default='***/BraTS2021/data')
parser.add_argument('--train_txt', type=str, default='***/BraTS2021/train.txt')
parser.add_argument('--valid_txt', type=str, default='***/BraTS2021/valid.txt')
parser.add_argument('--test_txt', type=str, default='***/BraTS2021/test.txt')
parser.add_argument('--train_log', type=str, default='results/UNet.txt')
parser.add_argument('--weights', type=str, default='results/UNet.pth')
parser.add_argument('--save_path', type=str, default='checkpoint/UNet')
args = parser.parse_args()
main(args)
训练集1000张,验证集125张,测试集126张。保存在验证集上损失最小的模型。
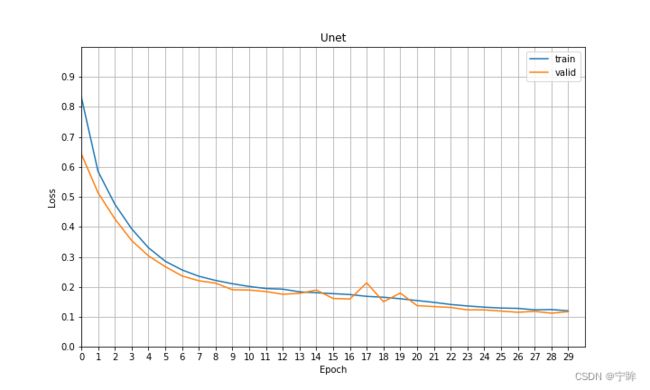
6.实验结果
训练30轮的loss曲线如上图所示,下面是我用不同的模型训练60轮,在测试集上的评价指标:
| 网络模型 | 三维数据大小 | ET | TC | WT | 均值 |
|---|---|---|---|---|---|
| UNet | 160×160×128 | 0.839 | 0.877 | 0.907 | 0.874 |
| Attention UNet | 160×160×128 | 0.850 | 0.877 | 0.915 | 0.881 |
| MCAUNet | 160×160×128 | 0.854 | 0.886 | 0.927 | 0.885 |
-
Attention UNet在UNet的基础上,在上采样模块引入像素注意力。
-
MCAUNet是我将CNN和Transformer结合,设计的一个网络。
确实,脑肿瘤分割相比其他三维分割任务,结果要好太多了,是一个非常适合练手的项目。感兴趣的同学可以按照我的步骤复现一下,效果也不会差。
代码我都放在上面了,码字不易,有用的话还请点个赞,后续也会更新图像分割和深度学习方面的内容,欢迎交流讨论。