【数据挖掘】主成分分析Python实现
目录
- 前言
- 数据预处理
-
- 导入库
- 读取数据
- 将五种癌症数据集合并
- 进行主成分分析
-
- 计算样本均值
- 计算样本协方差矩阵
- 计算特征值和特征向量
- 计算累计方差贡献率
- 特征向量选取
- 结果可视化
-
- 数据降维
- 降维前
- 降维后
- 写在最后
前言
是对一个数据挖掘作业的记录,数据集是老师提供的几种癌症的数据,我是直接在Jupyter中写的,中间会输出一些内容验证之类的
主要是参照这位写的:参考的大佬
数据预处理
导入库
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
读取数据
# 读取数据,查看数据的规模
BLCA_data = pd.read_csv(r'数据集/BLCA/rna.csv')
print('BLCA',BLCA_data.shape)
BRCA_data = pd.read_csv(r'数据集/BRCA/rna.csv')
print('BRCA',BRCA_data.shape)
KIRC_data = pd.read_csv(r'数据集/KIRC/rna.csv')
print('KIRC',KIRC_data.shape)
LUAD_data = pd.read_csv(r'数据集/LUAD/rna.csv')
print('LUAD',LUAD_data.shape)
PAAD_data = pd.read_csv(r'数据集/PAAD/rna.csv')
print('PAAD',PAAD_data.shape)
BLCA (3217, 400)
BRCA (3217, 1032)
KIRC (3217, 489)
LUAD (3217, 491)
PAAD (3217, 177)
# 查看其中一种类型癌症数据集的前6行
BLCA_data.head(6)
| gene_id | TCGA-HQ-A2OF | TCGA-GU-A767 | TCGA-ZF-AA4R | TCGA-DK-A1AC | TCGA-DK-A3IT | TCGA-GC-A3RD | TCGA-BT-A0YX | TCGA-FD-A6TE | TCGA-E7-A5KE | ... | TCGA-BT-A0S7 | TCGA-K4-A6FZ | TCGA-E5-A2PC | TCGA-DK-AA6Q | TCGA-XF-AAMY | TCGA-CU-A0YO | TCGA-E7-A7PW | TCGA-S5-A6DX | TCGA-GD-A76B | TCGA-ZF-AA4V | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | A2BP1|54715 | -0.402918 | -1.236959 | -1.453230 | -1.211589 | -1.288438 | -1.262086 | -1.033565 | -1.183245 | -1.076254 | ... | -1.282740 | -1.023637 | -0.366722 | -1.161285 | -1.380649 | -1.205136 | -0.875586 | -1.059148 | -1.393928 | -1.215641 |
| 1 | A2ML1|144568 | 0.717502 | 0.737891 | 1.584643 | 1.471932 | 0.414015 | 1.821084 | 2.515898 | 0.723830 | 0.668703 | ... | 0.163903 | 1.318229 | 0.879135 | 0.513603 | 1.571557 | 1.679199 | 1.222708 | -0.079802 | 1.480393 | 1.947340 |
| 2 | ACTL6B|51412 | -1.185185 | -1.403906 | -1.453230 | -1.211589 | -1.288438 | -1.262086 | -1.196075 | -1.183245 | -0.937518 | ... | -1.282740 | -1.291842 | -1.036778 | -1.161285 | -1.380649 | -1.368904 | -1.567590 | -1.425863 | -1.393928 | -1.215641 |
| 3 | ADAM6|8755 | 0.373826 | 1.411279 | 2.092282 | 3.186438 | 2.967457 | 1.437113 | 2.723775 | 2.320012 | 0.736963 | ... | 2.375413 | 1.964369 | 1.202000 | 1.824419 | 2.185051 | 2.175849 | 1.446352 | 3.591872 | 3.266818 | 2.514385 |
| 4 | ADAMDEC1|27299 | -0.489828 | -0.894004 | -0.205169 | 1.015179 | 0.089540 | 0.114903 | 0.664465 | 0.138955 | -1.076254 | ... | -0.408256 | -0.063113 | -0.147173 | 0.177490 | -0.254007 | 0.173888 | -0.951712 | 0.592750 | 0.260382 | 0.758897 |
| 5 | ALDH1A3|220 | 0.420170 | 0.563404 | 1.064257 | 0.460861 | 1.518204 | 1.809320 | 1.230912 | 1.488275 | 0.809302 | ... | 1.180098 | 1.751919 | 0.034835 | 1.512029 | 0.851459 | 1.645271 | 0.398217 | 0.599149 | -0.064256 | 1.339531 |
6 rows × 400 columns
# 查看另一种癌症数据集的前6行
BRCA_data.head(6)
| gene_id | TCGA-D8-A1XJ | TCGA-EW-A6SA | TCGA-D8-A27H | TCGA-E9-A247 | TCGA-AC-A2FG | TCGA-B6-A0RP | TCGA-BH-A0DH | TCGA-AN-A0AM | TCGA-D8-A1XZ | ... | TCGA-BH-A18G | TCGA-E2-A15J | TCGA-BH-A1F6 | TCGA-EW-A1P8 | TCGA-AO-A1KT | TCGA-A2-A0D0 | TCGA-3C-AALK | TCGA-AO-A1KS | TCGA-EW-A1OW | TCGA-OL-A5RU | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | A2BP1|54715 | -1.069946 | -1.075002 | -1.377040 | -1.307689 | -1.292405 | -1.231926 | -1.231272 | -1.396638 | -1.229635 | ... | -1.079587 | -1.018372 | -1.355431 | -1.156045 | -1.337455 | -1.189837 | -1.467466 | -1.339815 | -1.315667 | -1.421373 |
| 1 | A2ML1|144568 | 0.255748 | -0.877250 | 1.623317 | -1.307689 | -0.718106 | -1.135720 | -1.231272 | 0.253569 | 0.136528 | ... | -0.496468 | -1.135840 | 1.870474 | 0.995603 | -0.732074 | -0.500924 | -1.075625 | -0.708317 | 0.481910 | -0.781841 |
| 2 | ACTL6B|51412 | -1.253764 | -1.075002 | -1.377040 | -1.307689 | -1.458347 | -1.231926 | -1.231272 | -1.396638 | -1.229635 | ... | -1.079587 | -1.135840 | -1.200037 | -1.277135 | -1.337455 | -1.189837 | -1.467466 | -1.339815 | -1.162777 | -1.421373 |
| 3 | ADAM6|8755 | 1.949321 | 2.603630 | 2.016646 | 1.936839 | 2.851206 | 2.159315 | 3.494672 | 2.665196 | 3.116796 | ... | 1.503065 | 1.523513 | 3.238382 | 2.476680 | 2.039449 | 1.642455 | 2.529546 | 1.563575 | 2.519419 | 3.503925 |
| 4 | ADAMDEC1|27299 | -0.219076 | -0.343607 | -0.110169 | 0.410611 | -0.181777 | -0.254802 | -0.000519 | 0.218526 | 0.521942 | ... | -0.027635 | 0.073684 | 1.214923 | 0.885440 | -0.145622 | 0.466923 | 0.330690 | 0.485908 | 0.684071 | 0.723861 |
| 5 | ALDH1A3|220 | 1.138533 | 0.290696 | 2.239296 | 0.755849 | 1.327123 | 0.740471 | 1.101451 | 1.091598 | 0.884671 | ... | 2.251904 | 1.135105 | 1.978201 | 1.827946 | 0.834657 | 1.252129 | 1.159214 | 0.949791 | 1.215054 | 0.805760 |
6 rows × 1032 columns
经过上面的分析可以知道对于不同的数据集,它们的行数和行标签都是相同的,但列数和列标签都是不同的
所以对于这些数据集来说,行标签可以看作是数据的不同特征,而每一列则对应一个样本的数据
保留第一个数据集的第一列,然后将其与后面几个数据集除第一列之外的列进行合并
则:
BLCA:1-400
BRCA:401-1431
KIRC:1432-1919
LUAD:1920-2409
PAAD:2410-2585
将五种癌症数据集合并
# 将5种癌症数据集的除gene_id列之外的列进行合并
DataSet = pd.concat([BLCA_data,BRCA_data.iloc[:,1:],KIRC_data.iloc[:,1:],LUAD_data.iloc[:,1:],PAAD_data.iloc[:,1:]], axis=1)
DataSet.head(6)
| gene_id | TCGA-HQ-A2OF | TCGA-GU-A767 | TCGA-ZF-AA4R | TCGA-DK-A1AC | TCGA-DK-A3IT | TCGA-GC-A3RD | TCGA-BT-A0YX | TCGA-FD-A6TE | TCGA-E7-A5KE | ... | TCGA-FB-A7DR | TCGA-H8-A6C1 | TCGA-LB-A8F3 | TCGA-IB-7893 | TCGA-2J-AABF | TCGA-US-A77J | TCGA-F2-7273 | TCGA-HZ-A8P0 | TCGA-3A-A9IH | TCGA-2L-AAQA | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | A2BP1|54715 | -0.402918 | -1.236959 | -1.453230 | -1.211589 | -1.288438 | -1.262086 | -1.033565 | -1.183245 | -1.076254 | ... | -1.046787 | -1.561721 | -1.256692 | -1.345364 | -1.279792 | -1.621289 | -1.054519 | -1.455548 | -1.464518 | -1.471927 |
| 1 | A2ML1|144568 | 0.717502 | 0.737891 | 1.584643 | 1.471932 | 0.414015 | 1.821084 | 2.515898 | 0.723830 | 0.668703 | ... | -1.226226 | -0.680184 | -1.038366 | 1.252544 | -0.752025 | -1.621289 | -1.504779 | 0.680993 | 0.666947 | -0.915410 |
| 2 | ACTL6B|51412 | -1.185185 | -1.403906 | -1.453230 | -1.211589 | -1.288438 | -1.262086 | -1.196075 | -1.183245 | -0.937518 | ... | -1.126668 | -1.141486 | -0.768676 | -1.013273 | -0.542150 | -0.338250 | -0.368487 | -1.455548 | -0.454760 | -1.471927 |
| 3 | ADAM6|8755 | 0.373826 | 1.411279 | 2.092282 | 3.186438 | 2.967457 | 1.437113 | 2.723775 | 2.320012 | 0.736963 | ... | 2.500539 | 2.534327 | 1.895497 | 2.523738 | 2.771487 | 3.508540 | 2.503007 | 3.031195 | 2.018857 | 2.171331 |
| 4 | ADAMDEC1|27299 | -0.489828 | -0.894004 | -0.205169 | 1.015179 | 0.089540 | 0.114903 | 0.664465 | 0.138955 | -1.076254 | ... | 0.381645 | 0.286921 | -0.266635 | -0.161585 | 0.380947 | 0.772060 | 0.254188 | -0.122049 | -0.596158 | 0.098888 |
| 5 | ALDH1A3|220 | 0.420170 | 0.563404 | 1.064257 | 0.460861 | 1.518204 | 1.809320 | 1.230912 | 1.488275 | 0.809302 | ... | 1.530532 | 1.107393 | 0.618689 | 1.232431 | 1.062113 | 1.035272 | 1.220392 | 1.052504 | 0.985869 | 0.812195 |
6 rows × 2585 columns
对数据集进行转置,之后每一行对应一个样本数据,每一列表示一个样本特征
DataSet = DataSet.T
DataSet.head(6)
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 3207 | 3208 | 3209 | 3210 | 3211 | 3212 | 3213 | 3214 | 3215 | 3216 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| gene_id | A2BP1|54715 | A2ML1|144568 | ACTL6B|51412 | ADAM6|8755 | ADAMDEC1|27299 | ALDH1A3|220 | ATCAY|85300 | ATP10B|23120 | BCAS1|8537 | C10orf105|414152 | ... | SLC38A5|92745 | SLC44A3|126969 | SSX4|6759 | TMEM200B|399474 | TMEM26|219623 | TNFRSF10D|8793 | XAF1|54739 | YJEFN3|374887 | ZMAT1|84460 | ZNF415|55786 |
| TCGA-HQ-A2OF | -0.402918 | 0.717502 | -1.185185 | 0.373826 | -0.489828 | 0.42017 | -1.185185 | 1.841492 | 1.327818 | -0.301422 | ... | 0.275846 | 1.228001 | -0.959773 | -0.530559 | 0.637047 | 1.339706 | 0.520305 | 0.235093 | -0.383992 | 0.795605 |
| TCGA-GU-A767 | -1.236959 | 0.737891 | -1.403906 | 1.411279 | -0.894004 | 0.563404 | -1.236959 | 1.309958 | 2.015829 | 0.416162 | ... | 0.364331 | 1.484526 | -1.403906 | 0.689511 | -0.474266 | 0.378506 | 0.994647 | 0.776598 | 0.334363 | 0.577015 |
| TCGA-ZF-AA4R | -1.45323 | 1.584643 | -1.45323 | 2.092282 | -0.205169 | 1.064257 | -1.267946 | 0.207475 | 0.764633 | -0.786075 | ... | 1.350011 | 1.03154 | -1.45323 | 0.867536 | -0.145515 | 0.22999 | 1.210068 | 0.303078 | 0.91272 | 0.70952 |
| TCGA-DK-A1AC | -1.211589 | 1.471932 | -1.211589 | 3.186438 | 1.015179 | 0.460861 | -0.995834 | 1.576376 | 0.546906 | -0.855604 | ... | 0.637691 | 1.190068 | -0.348651 | 0.034641 | 0.030973 | 0.09397 | 1.089523 | 0.583813 | 0.503198 | 0.733853 |
| TCGA-DK-A3IT | -1.288438 | 0.414015 | -1.288438 | 2.967457 | 0.08954 | 1.518204 | -1.159105 | -0.035145 | 1.575849 | 0.220414 | ... | 0.598191 | 1.479815 | -1.288438 | 0.921235 | -0.3077 | 0.486069 | 1.311523 | 0.483139 | 0.262508 | 0.589681 |
6 rows × 3217 columns
# 此时第一行为特征标签,将非数据的这一行去掉
DataSet = DataSet.iloc[1:,:]
DataSet.iloc[:6,:]
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | ... | 3207 | 3208 | 3209 | 3210 | 3211 | 3212 | 3213 | 3214 | 3215 | 3216 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TCGA-HQ-A2OF | -0.402918 | 0.717502 | -1.185185 | 0.373826 | -0.489828 | 0.420170 | -1.185185 | 1.841492 | 1.327818 | -0.301422 | ... | 0.275846 | 1.228001 | -0.959773 | -0.530559 | 0.637047 | 1.339706 | 0.520305 | 0.235093 | -0.383992 | 0.795605 |
| TCGA-GU-A767 | -1.236959 | 0.737891 | -1.403906 | 1.411279 | -0.894004 | 0.563404 | -1.236959 | 1.309958 | 2.015829 | 0.416162 | ... | 0.364331 | 1.484526 | -1.403906 | 0.689511 | -0.474266 | 0.378506 | 0.994647 | 0.776598 | 0.334363 | 0.577015 |
| TCGA-ZF-AA4R | -1.453230 | 1.584643 | -1.453230 | 2.092282 | -0.205169 | 1.064257 | -1.267946 | 0.207475 | 0.764633 | -0.786075 | ... | 1.350011 | 1.031541 | -1.453230 | 0.867536 | -0.145515 | 0.229990 | 1.210068 | 0.303078 | 0.912720 | 0.709520 |
| TCGA-DK-A1AC | -1.211589 | 1.471932 | -1.211589 | 3.186439 | 1.015179 | 0.460861 | -0.995834 | 1.576376 | 0.546906 | -0.855604 | ... | 0.637691 | 1.190068 | -0.348651 | 0.034641 | 0.030973 | 0.093970 | 1.089523 | 0.583813 | 0.503198 | 0.733853 |
| TCGA-DK-A3IT | -1.288438 | 0.414015 | -1.288438 | 2.967457 | 0.089540 | 1.518204 | -1.159105 | -0.035145 | 1.575849 | 0.220414 | ... | 0.598191 | 1.479815 | -1.288438 | 0.921235 | -0.307700 | 0.486069 | 1.311522 | 0.483139 | 0.262508 | 0.589681 |
| TCGA-GC-A3RD | -1.262087 | 1.821084 | -1.262087 | 1.437113 | 0.114903 | 1.809320 | -0.993603 | 0.878451 | 2.058253 | 0.036055 | ... | 1.069556 | 1.383669 | -0.215558 | 0.234100 | -0.324477 | 0.379689 | 1.323748 | -0.026147 | 0.638123 | 0.096817 |
6 rows × 3217 columns
# 重新设置数据类型,此时DataSet为一个2584x3217的浮点数矩阵
DataSet = DataSet.astype('float32')
print(DataSet.shape)
(2584, 3217)
进行主成分分析
计算样本均值
Mean_vec = np.mean(DataSet, axis=0)
print('样本的均值:\n',Mean_vec)
样本的均值:
0 -1.161548
1 -0.474133
2 -1.251174
3 2.412248
4 0.266737
...
3212 0.521786
3213 1.303051
3214 0.133573
3215 0.774802
3216 0.601425
Length: 3217, dtype: float32
计算样本协方差矩阵
'''
# 纯数据运算
Cov_mat = (DataSet-Mean_vec).T.dot(DataSet-Mean_vec)/(DataSet.shape[0]-1)
print('协方差矩阵:\n',Cov_mat)
'''
# 使用numpy中计算协方差的函数
'''
协方差矩阵用于衡量两个变量之间相互依赖的程度
由于有3217个feature,故协方差矩阵的规模为3217x3217
'''
Cov_mat = np.cov(DataSet.T)
print('样本协方差矩阵:\n',Cov_mat)
Cov_mat.shape
样本协方差矩阵:
[[ 0.12676448 -0.02361515 0.02157561 ... 0.00519793 0.00904992
0.00673015]
[-0.02361515 1.00118482 -0.0053748 ... 0.07527039 -0.17714864
0.0090866 ]
[ 0.02157561 -0.0053748 0.13757093 ... 0.00533166 -0.00134267
0.00309703]
...
[ 0.00519793 0.07527039 0.00533166 ... 0.22596547 0.02969978
0.01703536]
[ 0.00904992 -0.17714864 -0.00134267 ... 0.02969978 0.20364439
0.04410373]
[ 0.00673015 0.0090866 0.00309703 ... 0.01703536 0.04410373
0.13098872]]
(3217, 3217)
计算特征值和特征向量
EigenValues, EigenVector = np.linalg.eig(Cov_mat)
# 数据特征量较大,这里计算结果为复数,统一取其实部
EigenValues = EigenValues.real
EigenVector = EigenVector.real
print('特征值:\n',EigenValues)
print('特征向量:\n',EigenVector)
特征值:
[ 2.31279350e+02 1.46770122e+02 8.81143724e+01 ... 2.86122668e-17
2.84073038e-17 -3.12152010e-17]
特征向量:
[[ 0.00030864 -0.00113702 -0.00123991 ... -0.00242919 -0.00277321
0.00190791]
[-0.02456387 0.02533867 -0.05603828 ... 0.00124872 0.00140361
-0.00075202]
[-0.00023562 -0.00033786 -0.00340553 ... -0.00034539 -0.00039286
0.00087827]
...
[-0.00238575 0.00450426 -0.01746374 ... -0.00422026 -0.00422903
0.00272333]
[ 0.00781593 -0.02092816 0.00775456 ... 0.01314091 0.01272775
0.01905482]
[-0.0030166 -0.01263667 -0.0111954 ... 0.02344654 0.03139416
0.01099676]]
# EigenPairs[i][0]表示一个特征值,EigenPairs[i][1]表示该特征值所对应的特征向量
EigenPairs = [(np.abs(EigenValues[i]), EigenVector[:,i]) for i in range(len(EigenValues))]
EigenPairs.sort(key=lambda x:x[0], reverse=True)
print('特征值降序排列:')
for i in EigenPairs:
print(i[0])
特征值降序排列:
231.2793503404339
146.77012152572306
88.11437236590861
......
1.6398199526831266e-18
1.4276145590349296e-18
1.4276145590349296e-18
计算累计方差贡献率
# 计算方差贡献率
tot = sum(EigenValues)
var_exp = [(i/tot)*100 for i in sorted(EigenValues, reverse=True)]
print(var_exp)
# 累计方差贡献率
Cum_var_exp = np.cumsum(var_exp)
Cum_var_exp
[18.501009256397158, 11.740760136661512, ... , -1.3125279499634018e-16, -2.0754934203007648e-16]
array([ 18.50100926, 30.24176939, 37.29040913, ..., 100., 100. , 100.])
特征向量选取
## 这里为方便作图,选取特征值较大的前两维的特征向量
Matrix = np.hstack((EigenPairs[0][1].reshape(DataSet.shape[1], 1),
EigenPairs[1][1].reshape(DataSet.shape[1], 1)))
print('Matrix:\n',Matrix)
Matrix:
[[ 0.00030864 -0.00113702]
[-0.02456387 0.02533867]
[-0.00023562 -0.00033786]
...
[-0.00238575 0.00450426]
[ 0.00781593 -0.02092816]
[-0.0030166 -0.01263667]]
结果可视化
数据降维
# tmp主要是将DaraSet转成array类型
HighDimSet = DataSet.iloc[:,:].values
# 将原数据矩阵乘以选取的特征向量进行降维
LowDimSet = HighDimSet.dot(Matrix)
LowDimSet
array([[ -2.78179211, 7.81840477],
[-13.39504631, 7.30085955],
[-16.8183728 , 3.87839756],
...,
[ -5.81835218, 8.30323684],
[ -8.52738732, 8.17363152],
[ -6.97177986, 11.89669388]])
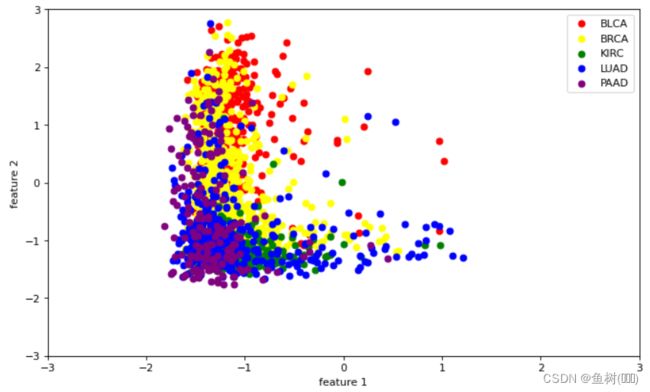
降维前
# LabelSet标识哪一行样本属于哪种癌症
LabelSet = ['BLCA']*399+['BRCA']*1031+['KIRC']*488+['LUAD']*490+['PAAD']*176
LabelSet = np.array(LabelSet)
LabelSet
array(['BLCA', 'BLCA', 'BLCA', ..., 'PAAD', 'PAAD', 'PAAD'], dtype='# 选取两种特征对不同的癌症种类进行区分
plt.figure(figsize=(10,6), dpi=80)
plt.xlim(-3,3)
plt.ylim(-3,3)
for lab,color in zip(('BLCA', 'BRCA', 'KIRC', 'LUAD', 'PAAD'),
('red','yellow','green','blue','purple')):
plt.scatter(HighDimSet[LabelSet==lab,0],
HighDimSet[LabelSet==lab,1],
label=lab,c=color)
plt.xlabel('feature 1')
plt.ylabel('feature 2')
plt.legend(loc='upper right')
plt.show()
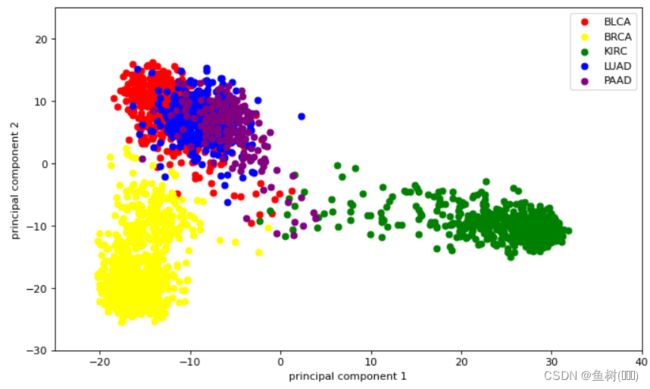
降维后
plt.figure(figsize=(10,6), dpi=80)
plt.xlim(-25,40)
plt.ylim(-30,25)
for lab,color in zip(('BLCA', 'BRCA', 'KIRC', 'LUAD', 'PAAD'),
('red','yellow','green','blue','purple')):
plt.scatter(LowDimSet[LabelSet==lab,0],
LowDimSet[LabelSet==lab,1],
label=lab,c=color)
plt.xlabel('principal component 1')
plt.ylabel('principal component 2')
plt.legend(loc='upper right')
plt.show()
写在最后
文章仅作记录,至于原理还有很多不懂的地方,结果我也不知道该是什么样的,把用这么多维特征来区分的事物降到两维来进行区分,我自己感觉已经很神奇了哈哈!