多目标粒子群优化算法 (MOPSO)(Matlab代码实现)
欢迎来到本博客❤️❤️❤️
博主优势:博客内容尽量做到思维缜密,逻辑清晰,为了方便读者。
⛳️座右铭:行百里者,半于九十。
目录
1 概述
2 运行结果
3 参考文献
4 Matlab代码实现
1 概述
多目标粒子群优化( Multi-objective Parti-cle Swarm Optimization,MOPSO)算法是由Carlos A、Coello C等在2004年提出来的,用于将单目标粒子群算法扩展应用于多目标优化问题中。MOPSO算法使用帕累托支配的概念来确定粒子的飞行方向,通过粒子间的支配关系比较得到非支配个体并存入全局精英库REP中,依据密度自适应网格估计法从全局精英库中选出全局最优解个体Gsest,同时依据新旧代的种群个体间的支配关系比较,来选出个体历史最优解Pest ,进而通过公式(6)来不断更新粒子群的速度和位置,最终用尽可能少的计算资源得到覆盖整个搜索空间、分布均匀、靠近真帕累托前沿的非劣解集。
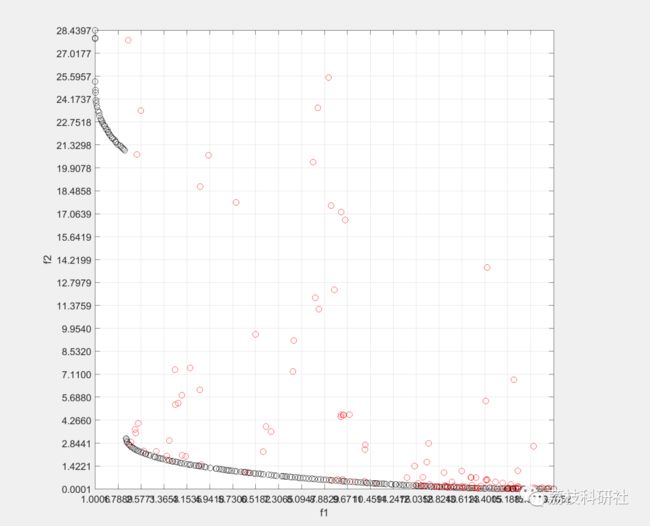
2 运行结果
部分代码:
% Plotting and verbose
if(size(POS_fit,2)==2)
h_fig = figure(1);
h_par = plot(POS_fit(:,1),POS_fit(:,2),'or'); hold on;
h_rep = plot(REP.pos_fit(:,1),REP.pos_fit(:,2),'ok'); hold on;
try
set(gca,'xtick',REP.hypercube_limits(:,1)','ytick',REP.hypercube_limits(:,2)');
axis([min(REP.hypercube_limits(:,1)) max(REP.hypercube_limits(:,1)) ...
min(REP.hypercube_limits(:,2)) max(REP.hypercube_limits(:,2))]);
grid on; xlabel('f1'); ylabel('f2');
end
drawnow;
end
if(size(POS_fit,2)==3)
h_fig = figure(1);
h_par = plot3(POS_fit(:,1),POS_fit(:,2),POS_fit(:,3),'or'); hold on;
h_rep = plot3(REP.pos_fit(:,1),REP.pos_fit(:,2),REP.pos_fit(:,3),'ok'); hold on;
try
set(gca,'xtick',REP.hypercube_limits(:,1)','ytick',REP.hypercube_limits(:,2)','ztick',REP.hypercube_limits(:,3)');
axis([min(REP.hypercube_limits(:,1)) max(REP.hypercube_limits(:,1)) ...
min(REP.hypercube_limits(:,2)) max(REP.hypercube_limits(:,2))]);
end
grid on; xlabel('f1'); ylabel('f2'); zlabel('f3');
drawnow;
axis square;
end
display(['Generation #0 - Repository size: ' num2str(size(REP.pos,1))]);
% Main MPSO loop
stopCondition = false;
while ~stopCondition
% Select leader
h = selectLeader(REP);
% Update speeds and positions
VEL = W.*VEL + C1*rand(Np,nVar).*(PBEST-POS) ...
+ C2*rand(Np,nVar).*(repmat(REP.pos(h,:),Np,1)-POS);
POS = POS + VEL;
% Perform mutation
POS = mutation(POS,gen,maxgen,Np,var_max,var_min,nVar,u_mut);
% Check boundaries
[POS,VEL] = checkBoundaries(POS,VEL,maxvel,var_max,var_min);
% Evaluate the population
POS_fit = fun(POS);
% Update the repository
REP = updateRepository(REP,POS,POS_fit,ngrid);
if(size(REP.pos,1)>Nr)
REP = deleteFromRepository(REP,size(REP.pos,1)-Nr,ngrid);
end
% Update the best positions found so far for each particle
pos_best = dominates(POS_fit, PBEST_fit);
best_pos = ~dominates(PBEST_fit, POS_fit);
best_pos(rand(Np,1)>=0.5) = 0;
if(sum(pos_best)>1)
PBEST_fit(pos_best,:) = POS_fit(pos_best,:);
PBEST(pos_best,:) = POS(pos_best,:);
end
if(sum(best_pos)>1)
PBEST_fit(best_pos,:) = POS_fit(best_pos,:);
PBEST(best_pos,:) = POS(best_pos,:);
end
% Plotting and verbose
if(size(POS_fit,2)==2)
figure(h_fig); delete(h_par); delete(h_rep);
h_par = plot(POS_fit(:,1),POS_fit(:,2),'or'); hold on;
h_rep = plot(REP.pos_fit(:,1),REP.pos_fit(:,2),'ok'); hold on;
try
set(gca,'xtick',REP.hypercube_limits(:,1)','ytick',REP.hypercube_limits(:,2)');
axis([min(REP.hypercube_limits(:,1)) max(REP.hypercube_limits(:,1)) ...
min(REP.hypercube_limits(:,2)) max(REP.hypercube_limits(:,2))]);
end
if(isfield(MultiObj,'truePF'))
try delete(h_pf); end
h_pf = plot(MultiObj.truePF(:,1),MultiObj.truePF(:,2),'.','color',0.8.*ones(1,3)); hold on;
end
grid on; xlabel('f1'); ylabel('f2');
drawnow;
axis square;
end
if(size(POS_fit,2)==3)
figure(h_fig); delete(h_par); delete(h_rep);
h_par = plot3(POS_fit(:,1),POS_fit(:,2),POS_fit(:,3),'or'); hold on;
h_rep = plot3(REP.pos_fit(:,1),REP.pos_fit(:,2),REP.pos_fit(:,3),'ok'); hold on;
try
set(gca,'xtick',REP.hypercube_limits(:,1)','ytick',REP.hypercube_limits(:,2)','ztick',REP.hypercube_limits(:,3)');
axis([min(REP.hypercube_limits(:,1)) max(REP.hypercube_limits(:,1)) ...
min(REP.hypercube_limits(:,2)) max(REP.hypercube_limits(:,2)) ...
min(REP.hypercube_limits(:,3)) max(REP.hypercube_limits(:,3))]);
end
if(isfield(MultiObj,'truePF'))
try delete(h_pf); end
h_pf = plot3(MultiObj.truePF(:,1),MultiObj.truePF(:,2),MultiObj.truePF(:,3),'.','color',0.8.*ones(1,3)); hold on;
end
grid on; xlabel('f1'); ylabel('f2'); zlabel('f3');
drawnow;
axis square;
end
display(['Generation #' num2str(gen) ' - Repository size: ' num2str(size(REP.pos,1))]);
% Update generation and check for termination
gen = gen + 1;
if(gen>maxgen), stopCondition = true; end
end
hold off;
end
% Function that updates the repository given a new population and its
% fitness
function REP = updateRepository(REP,POS,POS_fit,ngrid)
% Domination between particles
DOMINATED = checkDomination(POS_fit);
REP.pos = [REP.pos; POS(~DOMINATED,:)];
REP.pos_fit= [REP.pos_fit; POS_fit(~DOMINATED,:)];
% Domination between nondominated particles and the last repository
DOMINATED = checkDomination(REP.pos_fit);
REP.pos_fit= REP.pos_fit(~DOMINATED,:);
REP.pos = REP.pos(~DOMINATED,:);
% Updating the grid
REP = updateGrid(REP,ngrid);
end
% Function that corrects the positions and velocities of the particles that
% exceed the boundaries
function [POS,VEL] = checkBoundaries(POS,VEL,maxvel,var_max,var_min)
% Useful matrices
Np = size(POS,1);
MAXLIM = repmat(var_max(:)',Np,1);
MINLIM = repmat(var_min(:)',Np,1);
MAXVEL = repmat(maxvel(:)',Np,1);
MINVEL = repmat(-maxvel(:)',Np,1);
% Correct positions and velocities
VEL(VEL>MAXVEL) = MAXVEL(VEL>MAXVEL);
VEL(VEL
POS(POS>MAXLIM) = MAXLIM(POS>MAXLIM);
VEL(POS
% Function for checking the domination between the population. It
% returns a vector that indicates if each particle is dominated (1) or not
function dom_vector = checkDomination(fitness)
Np = size(fitness,1);
dom_vector = zeros(Np,1);
all_perm = nchoosek(1:Np,2); % Possible permutations
all_perm = [all_perm; [all_perm(:,2) all_perm(:,1)]];
d = dominates(fitness(all_perm(:,1),:),fitness(all_perm(:,2),:));
dominated_particles = unique(all_perm(d==1,2));
dom_vector(dominated_particles) = 1;
end
% Function that returns 1 if x dominates y and 0 otherwise
function d = dominates(x,y)
d = all(x<=y,2) & any(x
3 参考文献
References:
[1]Coello, C. A. C., Pulido, G. T., & Lechuga, M. S. (2004). Handling multiple objectives with particle swarm optimization. IEEE Tran- sactions on evolutionary computation, 8(3), 256-279.
[2]Sierra, M. R., & Coello, C. A. C. (2005, March). Improving PSO- based multi-objective optimization using crowding, mutation and -dominance. In International Conference on Evolutionary Multi-Criterion Optimization (pp. 505-519). Springer Berlin Heidelberg.
[3]刘俊贤,王宏强,陶新龙.基于改进多目标粒子群优化算法的雷达资源分配方法[J].中国电子科学研究院学报,2022,17(6):549-556565
[4]王万良,金雅文,陈嘉诚,李国庆,胡明志,董建杭.多角色多策略多目标粒子群优化算法[J].浙江大学学报:工学版,2022,56(3):531-541