geo差异表达分析_GEO数据库下载表达矩阵之后做下游分析
首先打开网页:
https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE104168
然后下载处理好的数据:
Excel打开:
然后就可以开始进行比较了:
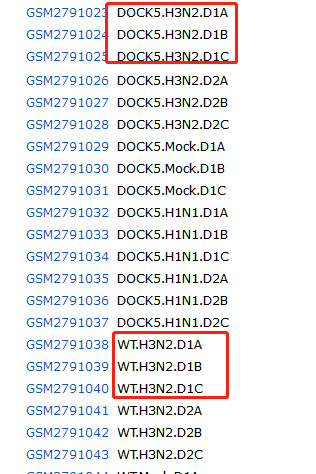
新建一个文件,并且从刚才的表达矩阵提取我们需要的几列数据:
本例选择:
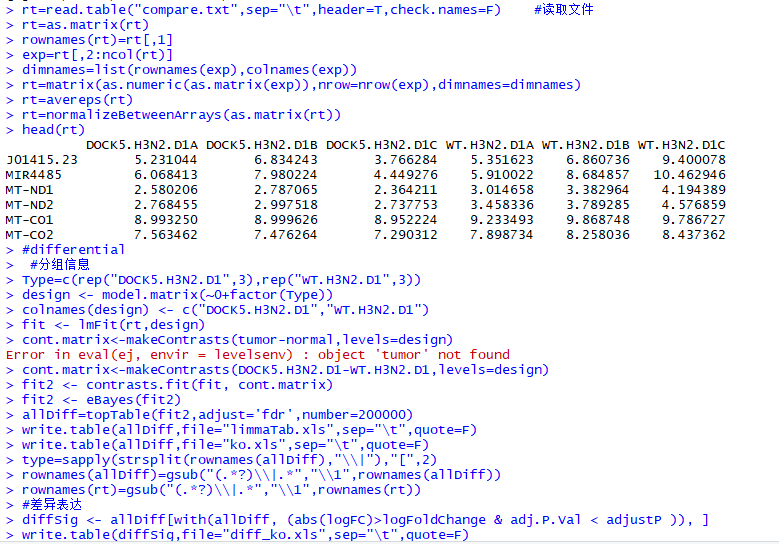
然后就在R进行操作:
#安装limma包,需要上网搜索最新的安装方式if (!requireNamespace("BiocManager", quietly = TRUE)) install.packages("BiocManager")BiocManager::install("limma")#导入包library("limma")library("impute")#设置参数logFoldChange=1adjustP=0.05getwd()setwd("") #设置工作目录rt=read.table("compare.txt",sep="\t",header=T,check.names=F) #读取文件rt=as.matrix(rt)rownames(rt)=rt[,1]exp=rt[,2:ncol(rt)]dimnames=list(rownames(exp),colnames(exp))rt=matrix(as.numeric(as.matrix(exp)),nrow=nrow(exp),dimnames=dimnames)rt=avereps(rt)rt=normalizeBetweenArrays(as.matrix(rt))#rt=log2(rt+1)#根据矩阵的值来确定,如果比较大则需要做log转换。#differential #分组信息Type=c(rep("DOCK5.H3N2.D1",3),rep("WT.H3N2.D1",3))design colnames(design) "DOCK5.H3N2.D1",fit cont.matrixfit2 fit2 allDiff=topTable(fit2,adjust='fdr',number=200000)write.table(allDiff,file="ko.xls",sep="\t",quote=F)type=sapply(strsplit(rownames(allDiff),"\\|"),"[",2)rownames(allDiff)=gsub("(.*?)\\|.*","\\1",rownames(allDiff))rownames(rt)=gsub("(.*?)\\|.*","\\1",rownames(rt))#差异表达diffSig logFoldChange & adj.P.Val < adjustP )), ]write.table(diffSig,file="diff_ko.xls",sep="\t",quote=F)#上调差异表达diffup logFoldChange & adj.P.Val < adjustP )), ]write.table(diffup,file="diff_ko_up.xls",sep="\t",quote=F)#下调差异表达diffdown write.table(diffdown,file="diff_ko_down.xls",sep="\t",quote=F)得到差异表达矩阵:
xMax=max(-log10(allDiff$adj.P.Val))yMax=max(abs(allDiff$logFC))plot(-log10(allDiff$adj.P.Val), allDiff$logFC, xlab="-log10(adj.P.Val)",ylab="logFC", main="Volcano", xlim=c(0,xMax),ylim=c(-yMax,yMax),yaxs="i",pch=20, cex=0.8)diffSub=subset(allDiff, adj.P.VallogFoldChange)points(-log10(diffSub$adj.P.Val), diffSub$logFC, pch=20, col="red",cex=0.8)diffSub=subset(allDiff, adj.P.Valpoints(-log10(diffSub$adj.P.Val), diffSub$logFC, pch=20, col="green",cex=0.8)abline(h=0,lty=2,lwd=3)#差异基因的表达水平,这是经过校正的hmExp=rt[rownames(diffSig),]diffExp=rbind(id=colnames(hmExp),hmExp)write.table(diffExp,file="heatmap.txt",sep="\t",quote=F,col.names=F)library(pheatmap)rt=read.table("heatmap.txt",sep="\t",header=T,row.names=1,check.names=F)Type=c(rep("DOCK5.H3N2.D1",3),rep("WT.H3N2.D1",3)) #修改对照和处理组样品数目fac=levels(factor(Type))rt1=rt[,Type==fac[1]]rt2=rt[,Type==fac[2]]rt=cbind(rt1,rt2)Type=c(rep(as.character(fac[1]),ncol(rt1)),rep(as.character(fac[2]),ncol(rt2)))names(Type)=colnames(rt)Type=as.data.frame(Type)tiff(file="heatmap.tiff", width = 30, #图片的宽度 height =30, #图片的高度 units ="cm", compression="lzw", bg="white", res=600)pheatmap(rt, annotation=Type, color = colorRampPalette(c("green", "black", "red"))(50), cluster_cols =F, fontsize = 8, fontsize_row=4, fontsize_col=8)dev.off()然后就可以搞其他的了:
将差异基因复制到deg.txt文件中。
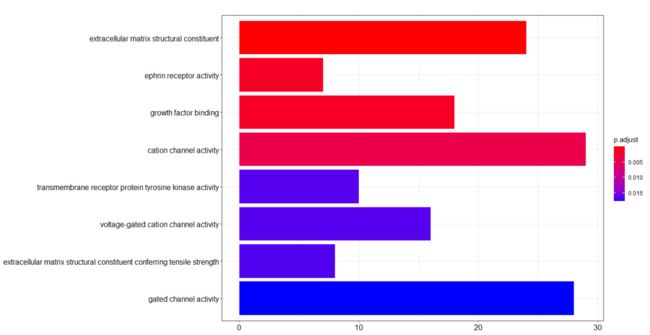
富集分析:
if (!requireNamespace("BiocManager", quietly = TRUE)) install.packages("BiocManager")BiocManager::install("topGo")BiocManager::install("clusterProfiler")BiocManager::install("org.Hs.eg.db")library(clusterProfiler)library(org.Hs.eg.db)library(topGO)getwd()a b eg = bitr(b, fromType="SYMBOL", toType="ENTREZID", OrgDb ="org.Hs.eg.db")data(geneList, package = "DOSE")gene gene ego gene = gene, keyType = "ENTREZID", OrgDb = org.Hs.eg.db, ont = "MF", pAdjustMethod = "BH", pvalueCutoff = 0.05, qvalueCutoff = 0.05, readable = TRUE)barplot(ego)ego gene = gene, keyType = "ENTREZID", OrgDb = org.Hs.eg.db, ont = "BP", pAdjustMethod = "BH", pvalueCutoff = 0.05, qvalueCutoff = 0.05, readable = TRUE)barplot(ego)ego gene = gene, keyType = "ENTREZID", OrgDb = org.Hs.eg.db, ont = "cc", pAdjustMethod = "BH", pvalueCutoff = 0.05, qvalueCutoff = 0.05, readable = TRUE)barplot(ego)dotplot(ego)ekegg gene = gene, keyType = "kegg", organism = 'hsa', pvalueCutoff = 0.05, pAdjustMethod = "BH", qvalueCutoff = 0.05)barplot(ekegg)dotplot(ekegg)网络分析:
STRING: functional protein association networks
https://string-db.org/
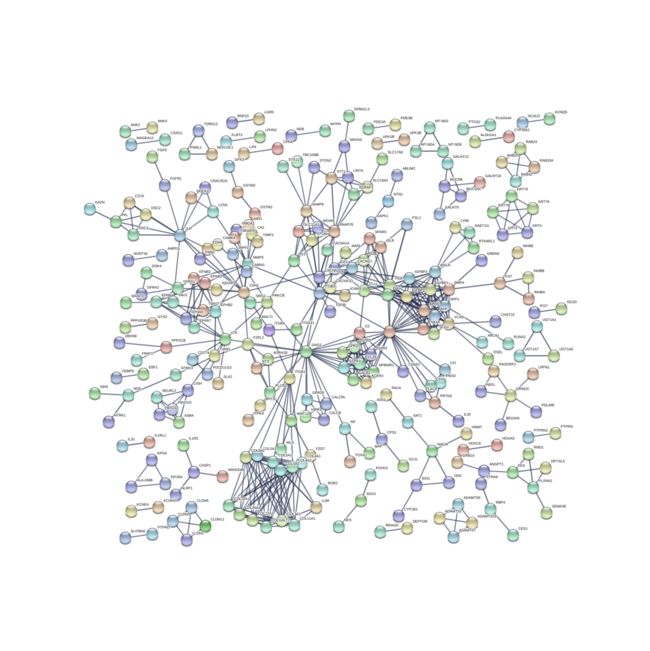
得到网络图:
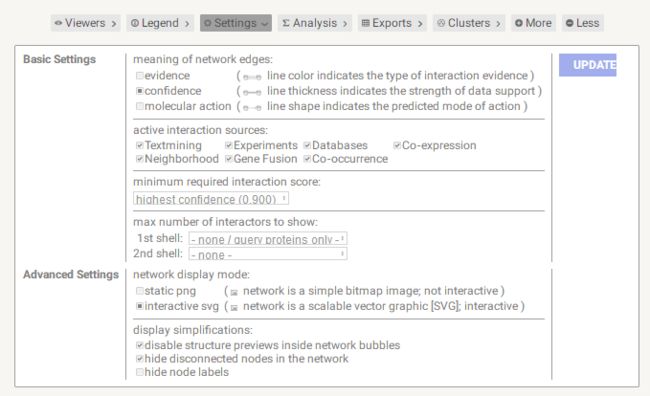
进行一下调节:
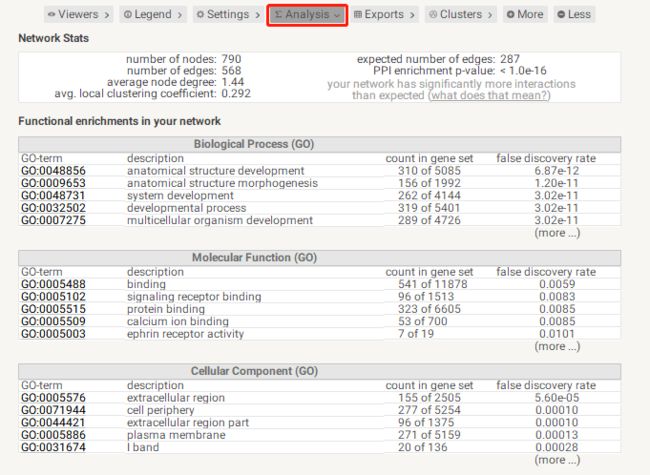
也有富集分析的结果:
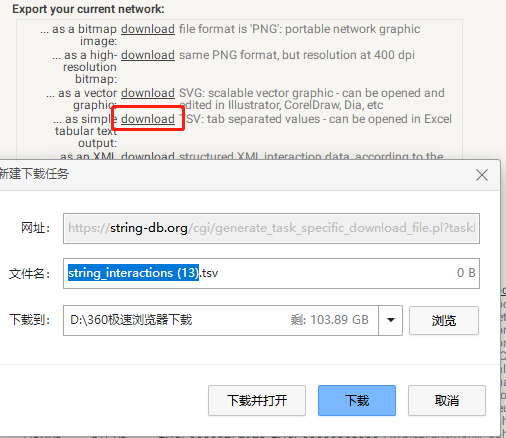
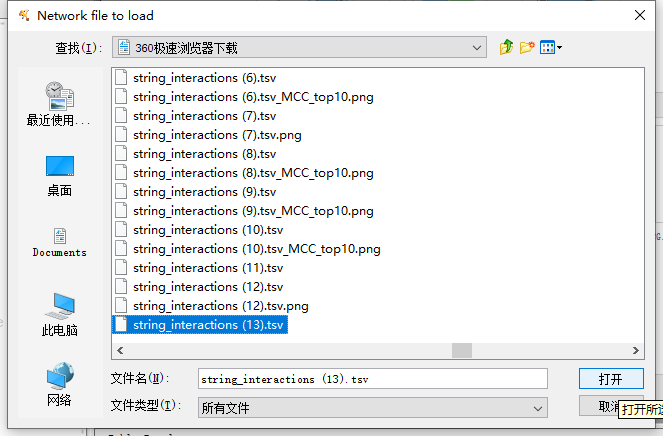
下载tsv文件:
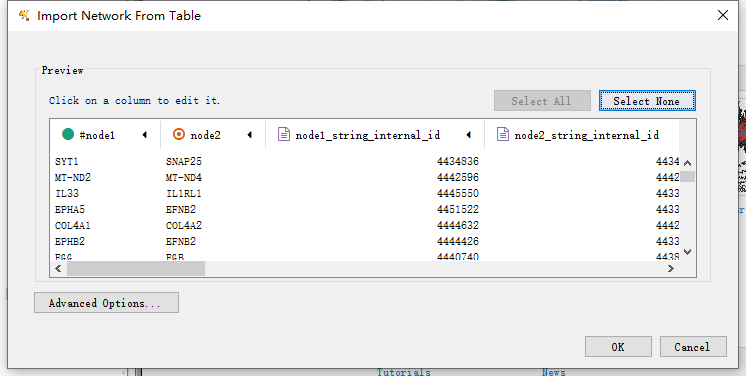
打开cytoscape,导入文件:
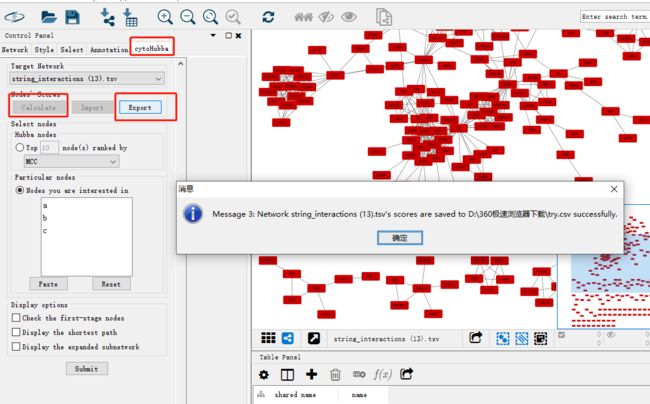
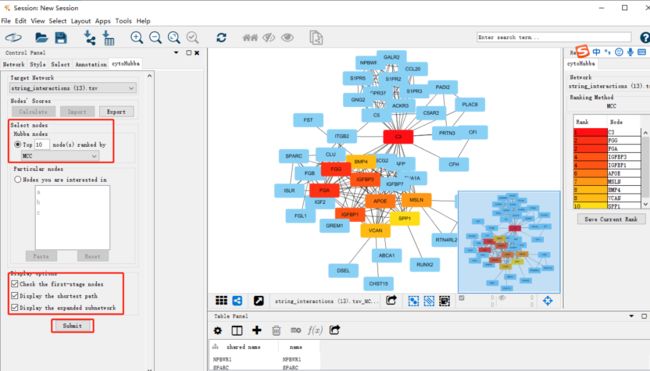
得到中心节点:
| Top 10 in network string_interactions (13).tsv ranked by MCC method | ||
| Rank | Name | Score |
| 1 | C3 | 6.23E+09 |
| 2 | FGG | 6.23E+09 |
| 2 | FGA | 6.23E+09 |
| 4 | IGFBP3 | 6.23E+09 |
| 4 | IGFBP1 | 6.23E+09 |
| 6 | APOE | 6.23E+09 |
| 7 | MSLN | 6.23E+09 |
| 8 | BMP4 | 6.23E+09 |
| 8 | VCAN | 6.23E+09 |
| 10 | SPP1 | 6.23E+09 |
参考资料:
不用编程的一篇SCI是怎么弄出来的?
https://mp.weixin.qq.com/s/0pmZ1EFHZDvVsZ4rdSv2Iw
从富集分析到统计学假设检验
https://mp.weixin.qq.com/s/3rermga6zgT7RWZyaAPn7A