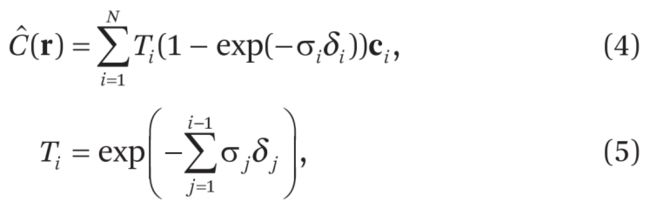mip-NeRF代码debug
代码:https://github.com/google/mipnerf
翻译解说:https://blog.csdn.net/qq_43620967/article/details/124458976
mip-NeRF-READNME
该存储库包含以下内容的代码版本 Mip-NeRF: A Multiscale Representation for Anti-Aliasing Neural Radiance Fields. 这个实现是用JAX写的,是Google的JaxNeRF实现的一个分支。如果您遇到任何问题,请联系Jon Barron

Abstract
Installation
我们建议使用 Anaconda 来建立环境。运行以下命令:
# Clone the repo
git clone https://github.com/google/mipnerf.git; cd mipnerf
# Create a conda environment, note you can use python 3.6-3.8 as
# one of the dependencies (TensorFlow) hasn't supported python 3.9 yet.
conda create --name mipnerf python=3.6.13; conda activate mipnerf
# Prepare pip
conda install pip; pip install --upgrade pip
# Install requirements
pip install -r requirements.txt
pip install -r requirements.txt会报错
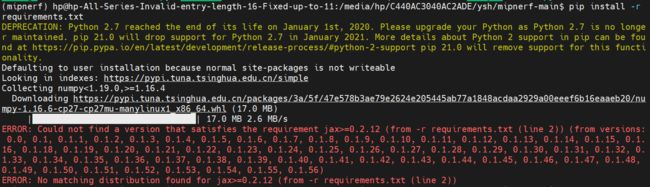
改成
pip3 install -r requirements.txt
pip3 install -r requirements.txt --default-timeout=500
[Optional] Install GPU and TPU support for Jax
# Remember to change cuda101 to your CUDA version, e.g. cuda110 for CUDA 11.0.
pip install --upgrade jax jaxlib==0.1.65+cuda101 -f https://storage.googleapis.com/jax-releases/jax_releases.html
我安装的是
pip3 install --upgrade jax==0.2.3 jaxlib==0.1.69+cuda111 -f https://storage.googleapis.com/jax-releases/jax_releases.html
Data
然后,你需要从NeRF官方Google Drive下载数据集。请下载并解压nerf_synthetic.zip和nerf_llff_data.zip.
Generate multiscale dataset
您可以通过运行以下命令来生成本文中使用的多尺度数据集,
python scripts/convert_blender_data.py --blenderdir /nerf_synthetic --outdir /multiscale
Running
在本文中使用的三个数据集的单个场景上训练mip-NeRF的示例脚本可以在scripts/中找到。您需要将路径更改为指向数据集所在的位置。我们的模型和一些消融的Gin配置文件可以在configs/中找到。在scripts/中可以找到对每个场景的测试集进行评估的示例脚本,之后您可以使用scripts/summary . ipynb来生成所有场景的错误度量,其格式与本文表格中使用的格式相同。
OOM errors
您可能需要减小批处理大小,以避免内存不足错误。例如,该模型可以使用以下标志在NVIDIA 3080 (10Gb)上运行.
--gin_param="Config.batch_size = 1024"
Citation
If you use this software package, please cite our paper:
@misc{barron2021mipnerf,
title={Mip-NeRF: A Multiscale Representation for Anti-Aliasing Neural Radiance Fields},
author={Jonathan T. Barron and Ben Mildenhall and Matthew Tancik and Peter Hedman and Ricardo Martin-Brualla and Pratul P. Srinivasan},
year={2021},
eprint={2103.13415},
archivePrefix={arXiv},
primaryClass={cs.CV}
}
Acknowledgements
Thanks to Boyang Deng for JaxNeRF.
项目地址
/media/hp/C440AC3040AC2ADE/ysh/mipnerf-main/
scripts/convert_blender_data.py
def main
def main(unused_argv):
blenderdir = FLAGS.blenderdir
outdir = FLAGS.outdir
n_down = FLAGS.n_down
if not os.path.exists(outdir):
os.makedirs(outdir)
dirs = [os.path.join(blenderdir, f) for f in os.listdir(blenderdir)]
dirs = [d for d in dirs if os.path.isdir(d)]
print(dirs)
for basedir in dirs:
print()
newdir = os.path.join(outdir, os.path.basename(basedir))
print('Converting from', basedir, 'to', newdir)
convert_to_nerfdata(basedir, newdir, n_down)
def load_renderings
def load_renderings(data_dir, split):
"""Load images and metadata from disk."""
f = 'transforms_{}.json'.format(split)
with open(path.join(data_dir, f), 'r') as fp:
meta = json.load(fp)
images = []
cams = []
print('Loading imgs')
for frame in meta['frames']:
fname = os.path.join(data_dir, frame['file_path'] + '.png')
with open(fname, 'rb') as imgin:
image = np.array(Image.open(imgin), dtype=np.float32) / 255.
cams.append(frame['transform_matrix'])
images.append(image)
ret = {}
ret['images'] = np.stack(images, axis=0)
print('Loaded all images, shape is', ret['images'].shape)
ret['camtoworlds'] = np.stack(cams, axis=0)
w = ret['images'].shape[2]
camera_angle_x = float(meta['camera_angle_x'])
ret['focal'] = .5 * w / np.tan(.5 * camera_angle_x)
return ret
transforms_train.json
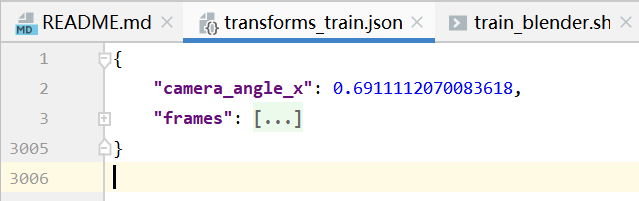
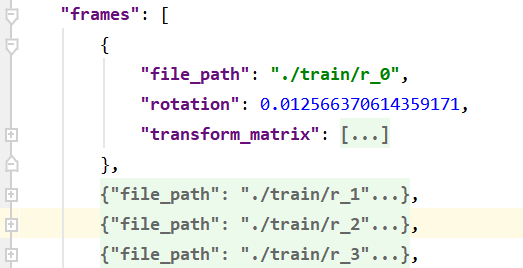
camera_angle_x 用来计算focal ,是相机的水平视场
ret['focal'] = .5 * w / np.tan(.5 * camera_angle_x)
file_path 用来获取图片路径
fname = os.path.join(data_dir, frame['file_path'] + '.png')
rotation 没用到
transform_matrix 用作相机坐标到世界坐标的转换
是从相机坐标到世界坐标转换的姿态矩阵(camera-to-world) 即

相机坐标系=Cex * 世界坐标系
Cex逆 * 相机坐标系=世界坐标系
cams.append(frame['transform_matrix'])
ret['camtoworlds'] = np.stack(cams, axis=0)
def convert_to_nerfdata
def convert_to_nerfdata(basedir, newdir, n_down):
"""Convert Blender data to multiscale."""
if not os.path.exists(newdir):
os.makedirs(newdir)
splits = ['train', 'val', 'test']
bigmeta = {}
# Foreach split in the dataset
for split in splits:
print('Split', split)
# Load everything
data = load_renderings(basedir, split)
# Save out all the images
imgdir = 'images_{}'.format(split)
os.makedirs(os.path.join(newdir, imgdir), exist_ok=True)
fnames = []
widths = []
heights = []
focals = []
cam2worlds = []
lossmults = []
labels = []
nears, fars = [], []
f = data['focal']
print('Saving images')
for i, img in enumerate(data['images']):
for j in range(n_down):
fname = '{}/{:03d}_d{}.png'.format(imgdir, i, j)
fnames.append(fname)
fname = os.path.join(newdir, fname)
with open(fname, 'wb') as imgout:
img8 = Image.fromarray(np.uint8(img * 255))
img8.save(imgout)
widths.append(img.shape[1])
heights.append(img.shape[0])
focals.append(f / 2**j)
cam2worlds.append(data['camtoworlds'][i].tolist())
lossmults.append(4.**j)
labels.append(j)
nears.append(2.)
fars.append(6.)
img = down2(img)
def down2(img):
sh = img.shape
return np.mean(np.reshape(img, [sh[0] // 2, 2, sh[1] // 2, 2, -1]), (1, 3))
图片大小一路缩小
sh变化:
(800, 800, 4)->(100, 100, 4)
shape 值变化:
[800, 400, 200, 100]
# Create metadata
meta = {}
meta['file_path'] = fnames
meta['cam2world'] = cam2worlds
meta['width'] = widths
meta['height'] = heights
meta['focal'] = focals
meta['label'] = labels
meta['near'] = nears
meta['far'] = fars
meta['lossmult'] = lossmults
fx = np.array(focals)
fy = np.array(focals)
cx = np.array(meta['width']) * .5
cy = np.array(meta['height']) * .5
arr0 = np.zeros_like(cx)
arr1 = np.ones_like(cx)
k_inv = np.array([
[arr1 / fx, arr0, -cx / fx],
[arr0, -arr1 / fy, cy / fy],
[arr0, arr0, -arr1],
])
k_inv = np.moveaxis(k_inv, -1, 0)
meta['pix2cam'] = k_inv.tolist()
bigmeta[split] = meta
for k in bigmeta:
for j in bigmeta[k]:
print(k, j, type(bigmeta[k][j]), np.array(bigmeta[k][j]).shape)
jsonfile = os.path.join(newdir, 'metadata.json')
with open(jsonfile, 'w') as f:
json.dump(bigmeta, f, ensure_ascii=False, indent=4)

meta[‘pix2cam’]

二维图片的坐标 和 相机坐标系中的坐标 存在下面的转换关系: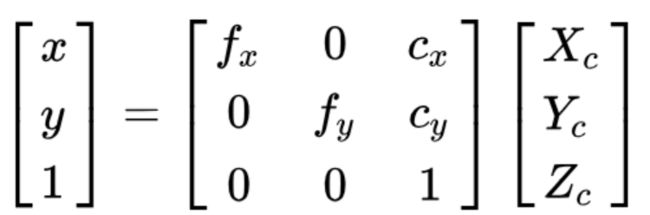
其中,矩阵 指的是相机的内参,包含焦距 (focal length) fx 以及图像中心点的坐标 cx 。
k_inv 是 相机的内参矩阵的逆 即 pix2cam 图像到相机矩阵
对于相同的数据集,相机的内参矩阵一般是固定的,一般会在一个叫 intrinsics.txt 的文件里面给出。
train_blender.sh
# Script for training on the Blender dataset.
SCENE=lego
EXPERIMENT=debug
TRAIN_DIR=/Users/barron/tmp/nerf_results/$EXPERIMENT/$SCENE
DATA_DIR=/Users/barron/data/nerf_synthetic/$SCENE
rm $TRAIN_DIR/*
python -m train \
--data_dir=$DATA_DIR \
--train_dir=$TRAIN_DIR \
--gin_file=configs/blender.gin \
--logtostderr
python -m
将库中的python模块用作脚本去运行
1.python xxx.py
2.python -m xxx.py
这是两种加载py文件的方式:
1叫做直接运行
2相当于import,叫做当做模块来启动
不同的加载py文件的方式,主要是影响sys.path这个属性。sys.path相当于Linux中的PATH。
这就是当前Python解析器运行的环境,Python解析器会在这些目录下去寻找依赖库。
运行脚本
将目录改成自己的数据集地址
TRAIN_DIR=/media/hp/C440AC3040AC2ADE/ysh/mipnerf-main/nerf_example_data/nerf_results/$EXPERIMENT/$SCENE
DATA_DIR=/media/hp/C440AC3040AC2ADE/ysh/mipnerf-main/nerf_example_data/nerf_synthetic/$SCENE
conda activate mipnerf
bash scripts/train_blender.sh
configs/blender.gin
Config.dataset_loader = 'blender'
Config.batching = 'single_image'
train.py
from absl import app
from absl import flags
from internal import datasets
from internal import math
from internal import models
from internal import utils
from internal import vis
config配置
FLAGS 作用及用法
FLAGS = flags.FLAGS
utils.define_common_flags()
flags.DEFINE_integer('render_every', 5000,
'The number of steps between test set image renderings.')#测试集图像渲染之间的步骤数
jax.config.parse_flags_with_absl()
utils.define_common_flags()
def define_common_flags():
# Define the flags used by both train.py and eval.py
flags.DEFINE_multi_string('gin_file', None,
'List of paths to the config files.')
flags.DEFINE_multi_string(
'gin_param', None, 'Newline separated list of Gin parameter bindings.') #换行符分隔的Gin参数绑定列表
flags.DEFINE_string('train_dir', None, 'where to store ckpts and logs')
flags.DEFINE_string('data_dir', None, 'input data directory.')
flags.DEFINE_integer(
'chunk', 8192,
'the size of chunks for evaluation inferences, set to the value that'
'fits your GPU/TPU memory.')
main
1.config
def main(unused_argv):
rng = random.PRNGKey(20200823)
# Shift the numpy random seed by host_id() to shuffle data loaded by different hosts.
np.random.seed(20201473 + jax.host_id())
config = utils.load_config()
if config.batch_size % jax.device_count() != 0:
raise ValueError('Batch size must be divisible by the number of devices.')
utils.load_config()
def load_config():
gin.parse_config_files_and_bindings(flags.FLAGS.gin_file,
flags.FLAGS.gin_param)
return Config()
class Config
class Config:
"""Configuration flags for everything."""
dataset_loader: str = 'multicam' # The type of dataset loader to use.
batching: str = 'all_images' # Batch composition, [single_image, all_images].
batch_size: int = 4096 # The number of rays/pixels in each batch.
factor: int = 0 # The downsample factor of images, 0 for no downsampling.
spherify: bool = False # Set to True for spherical 360 scenes.
render_path: bool = False # If True, render a path. Used only by LLFF.
llffhold: int = 8 # Use every Nth image for the test set. Used only by LLFF.
lr_init: float = 5e-4 # The initial learning rate.
lr_final: float = 5e-6 # The final learning rate.
lr_delay_steps: int = 2500 # The number of "warmup" learning steps.
lr_delay_mult: float = 0.01 # How much sever the "warmup" should be.
grad_max_norm: float = 0. # Gradient clipping magnitude, disabled if == 0.
grad_max_val: float = 0. # Gradient clipping value, disabled if == 0.
max_steps: int = 1000000 # The number of optimization steps.
save_every: int = 100000 # The number of steps to save a checkpoint.
print_every: int = 100 # The number of steps between reports to tensorboard.
gc_every: int = 10000 # The number of steps between garbage collections.
test_render_interval: int = 1 # The interval between images saved to disk.
disable_multiscale_loss: bool = False # If True, disable multiscale loss.
randomized: bool = True # Use randomized stratified sampling.
near: float = 2. # Near plane distance.
far: float = 6. # Far plane distance.
coarse_loss_mult: float = 0.1 # How much to downweight the coarse loss(es).
weight_decay_mult: float = 0. # The multiplier on weight decay.
white_bkgd: bool = True # If True, use white as the background (black o.w.).
2.dataset
dataset = datasets.get_dataset('train', FLAGS.data_dir, config)
test_dataset = datasets.get_dataset('test', FLAGS.data_dir, config)
def get_dataset
def get_dataset(split, train_dir, config):
return dataset_dict[config.dataset_loader](split, train_dir, config)
dataset_dict = {
'blender': Blender,
'llff': LLFF,
'multicam': Multicam,
}
class Blender
class Blender(Dataset):
"""Blender Dataset."""
class Dataset
class Dataset(threading.Thread):
"""Dataset Base Class."""
def __init__(self, split, data_dir, config):
super(Dataset, self).__init__()
self.queue = queue.Queue(3) # Set prefetch buffer to 3 batches.
self.daemon = True
self.split = split
self.data_dir = data_dir
self.near = config.near
self.far = config.far
if split == 'train':
self._train_init(config)
elif split == 'test':
self._test_init(config)
else:
raise ValueError(
'the split argument should be either \'train\' or \'test\', set'
'to {} here.'.format(split))
self.batch_size = config.batch_size // jax.host_count()
self.batching = config.batching
self.render_path = config.render_path
self.start()
def _train_init
def _train_init(self, config):
"""Initialize training."""
self._load_renderings(config) #加载图片 以及 对应图片相机信息
self._generate_rays() #生成光线
if config.batching == 'all_images':
# flatten the ray and image dimension together.
self.images = self.images.reshape([-1, 3])
self.rays = utils.namedtuple_map(lambda r: r.reshape([-1, r.shape[-1]]),
self.rays)
elif config.batching == 'single_image':
self.images = self.images.reshape([-1, self.resolution, 3])
self.rays = utils.namedtuple_map(
lambda r: r.reshape([-1, self.resolution, r.shape[-1]]), self.rays)
else:
raise NotImplementedError(
f'{config.batching} batching strategy is not implemented.')
def _generate_rays
描述了光线的具体生成方法,一条光线包含以下内容
self.rays = utils.Rays(
origins=origins,
directions=directions,
viewdirs=viewdirs,
radii=radii,
lossmult=ones,
near=ones * self.near,
far=ones * self.far)
# TODO(bydeng): Swap this function with a more flexible camera model.
def _generate_rays(self):
"""Generating rays for all images."""
x, y = np.meshgrid( # pylint: disable=unbalanced-tuple-unpacking
np.arange(self.w, dtype=np.float32), # X-Axis (columns)
np.arange(self.h, dtype=np.float32), # Y-Axis (rows)
indexing='xy')
camera_dirs = np.stack(
[(x - self.w * 0.5 + 0.5) / self.focal,
-(y - self.h * 0.5 + 0.5) / self.focal, -np.ones_like(x)],
axis=-1)
directions = ((camera_dirs[None, ..., None, :] *
self.camtoworlds[:, None, None, :3, :3]).sum(axis=-1))
origins = np.broadcast_to(self.camtoworlds[:, None, None, :3, -1],
directions.shape)
viewdirs = directions / np.linalg.norm(directions, axis=-1, keepdims=True)
# Distance from each unit-norm direction vector to its x-axis neighbor.
dx = np.sqrt(
np.sum((directions[:, :-1, :, :] - directions[:, 1:, :, :])**2, -1))
dx = np.concatenate([dx, dx[:, -2:-1, :]], 1)
# Cut the distance in half, and then round it out so that it's
# halfway between inscribed by / circumscribed about the pixel.
radii = dx[..., None] * 2 / np.sqrt(12)
ones = np.ones_like(origins[..., :1])
self.rays = utils.Rays(
origins=origins,
directions=directions,
viewdirs=viewdirs,
radii=radii,
lossmult=ones,
near=ones * self.near,
far=ones * self.far)
utils.Rays
Rays = collections.namedtuple(
'Rays',
('origins', 'directions', 'viewdirs', 'radii', 'lossmult', 'near', 'far'))
3.model
rng, key = random.split(rng)
model, variables = models.construct_mipnerf(key, dataset.peek())
num_params = jax.tree_util.tree_reduce(
lambda x, y: x + jnp.prod(jnp.array(y.shape)), variables, initializer=0)
print(f'Number of parameters being optimized: {num_params}')
optimizer = flax.optim.Adam(config.lr_init).create(variables)
state = utils.TrainState(optimizer=optimizer)
del optimizer, variables
def peek(self):
"""在不出列的情况下,查看下一批训练或测试示例.
Returns:
batch: dict, has 'pixels' and 'rays'.
"""
x = self.queue.queue[0].copy() # Make a copy of the front of the queue.
if self.split == 'train':
return utils.shard(x)
else:
return utils.to_device(x)
def shard(xs):
"""沿着第一维将多个设备的数据分割成碎片。"""
return jax.tree_map(
lambda x: x.reshape((jax.local_device_count(), -1) + x.shape[1:]), xs)
def construct_mipnerf
def construct_mipnerf(rng, example_batch):
"""Construct a Neural Radiance Field.
Args:
rng: jnp.ndarray. 随机数生成器。
example_batch: dict, an example of a batch of data.
Returns:
model: nn.Model. Nerf model with parameters.
state: flax.Module.state. 有状态参数的Nerf模型状态.
"""
model = MipNerfModel()
key, rng = random.split(rng)
init_variables = model.init(
key,
rng=rng,
rays=utils.namedtuple_map(lambda x: x[0], example_batch['rays']),
randomized=False,
white_bkgd=False)
return model, init_variables
model值
MipNerfModel(
# attributes
num_samples = 128
num_levels = 2
resample_padding = 0.01
stop_level_grad = True
use_viewdirs = True
lindisp = False
ray_shape = 'cone'
min_deg_point = 0
max_deg_point = 16
deg_view = 4
density_activation = softplus
density_noise = 0.0
density_bias = -1.0
rgb_activation = sigmoid
rgb_padding = 0.001
disable_integration = False
)
4.学习率
learning_rate_fn = functools.partial(
math.learning_rate_decay,
lr_init=config.lr_init,
lr_final=config.lr_final,
max_steps=config.max_steps,
lr_delay_steps=config.lr_delay_steps,
lr_delay_mult=config.lr_delay_mult)
math.learning_rate_decay
连续学习率衰减函数
def learning_rate_decay(step,
lr_init,
lr_final,
max_steps,
lr_delay_steps=0,
lr_delay_mult=1):
"""Continuous learning rate decay function.
当步长=0时,返回的速率为lr_init,当步长=max_steps时,返回的速率为lr_final,
并且 在别处是对数线性插值的(相当于指数衰减)。
如果lr_delay_steps>0,那么学习速率将由lr_delay_mult的某个平滑函数来缩放,
使得初始学习速率在优化开始时是lr_init*lr_delay_mult,
但是当steps>lr_delay_steps时将被缓和回到正常学习速率。
Args:
step: int, the current optimization step.
lr_init: float, the initial learning rate.
lr_final: float, the final learning rate.
max_steps: int, the number of steps during optimization.
lr_delay_steps: int, the number of steps to delay the full learning rate.
lr_delay_mult: float, the multiplier on the rate when delaying it.
Returns:
lr: the learning for current step 'step'.
"""
if lr_delay_steps > 0:
# A kind of reverse cosine decay.
delay_rate = lr_delay_mult + (1 - lr_delay_mult) * jnp.sin(
0.5 * jnp.pi * jnp.clip(step / lr_delay_steps, 0, 1))
else:
delay_rate = 1.
t = jnp.clip(step / max_steps, 0, 1)
log_lerp = jnp.exp(jnp.log(lr_init) * (1 - t) + jnp.log(lr_final) * t)
return delay_rate * log_lerp
5.函数映射
train_pstep = jax.pmap(
functools.partial(train_step, model, config),
axis_name='batch',
in_axes=(0, 0, 0, None),
donate_argnums=(2,))
render_eval_pfn = jax.pmap(
render_eval_fn,
in_axes=(None, None, 0), # Only distribute the data input.
donate_argnums=(2,),
axis_name='batch',
)
ssim_fn = jax.jit(functools.partial(math.compute_ssim, max_val=1.))
6.加载已训练模型
if not utils.isdir(FLAGS.train_dir):
utils.makedirs(FLAGS.train_dir)
state = checkpoints.restore_checkpoint(FLAGS.train_dir, state)
# Resume training a the step of the last checkpoint.
init_step = state.optimizer.state.step + 1
state = flax.jax_utils.replicate(state)
7.summary_writer
if jax.host_id() == 0:
summary_writer = tensorboard.SummaryWriter(FLAGS.train_dir)
8.迭代
# Prefetch_buffer_size = 3 x batch_size 预缓冲区大小
pdataset = flax.jax_utils.prefetch_to_device(dataset, 3)
rng = rng + jax.host_id() # Make random seed separate across hosts.使随机种子在主机之间分离。
keys = random.split(rng, jax.local_device_count()) # For pmapping RNG keys.
gc.disable() # Disable automatic garbage collection for efficiency.禁用自动垃圾收集以提高效率。
stats_trace = []
reset_timer = True
for step, batch in zip(range(init_step, config.max_steps + 1), pdataset):
if reset_timer:
t_loop_start = time.time()
reset_timer = False
lr = learning_rate_fn(step)
state, stats, keys = train_pstep(keys, state, batch, lr)
if jax.host_id() == 0:
stats_trace.append(stats)
if step % config.gc_every == 0: # The number of steps between garbage collections.
gc.collect() #如果没有参数,运行完整的收集。可选参数可以是指定要收集哪generation的整数。如果generation无效,将引发ValueError。
Log training summaries
这被放在host_id检查之后,因为在多主机评估中,所有主机都需要运行推理,即使我们只使用host 0来记录结果。
# Log training summaries. This is put behind a host_id check because in multi-host evaluation, all hosts need to run inference even though we only use host 0 to record results.
if jax.host_id() == 0:
if step % config.print_every == 0:
summary_writer.scalar('num_params', num_params, step)
summary_writer.scalar('train_loss', stats.loss[0], step)
summary_writer.scalar('train_psnr', stats.psnr[0], step)
for i, l in enumerate(stats.losses[0]):
summary_writer.scalar(f'train_losses_{i}', l, step)
for i, p in enumerate(stats.psnrs[0]):
summary_writer.scalar(f'train_psnrs_{i}', p, step)
summary_writer.scalar('weight_l2', stats.weight_l2[0], step)
avg_loss = np.mean(np.concatenate([s.loss for s in stats_trace]))
avg_psnr = np.mean(np.concatenate([s.psnr for s in stats_trace]))
max_grad_norm = np.max(
np.concatenate([s.grad_norm for s in stats_trace]))
avg_grad_norm = np.mean(
np.concatenate([s.grad_norm for s in stats_trace]))
max_clipped_grad_norm = np.max(
np.concatenate([s.grad_norm_clipped for s in stats_trace]))
max_grad_max = np.max(
np.concatenate([s.grad_abs_max for s in stats_trace]))
stats_trace = []
summary_writer.scalar('train_avg_loss', avg_loss, step)
summary_writer.scalar('train_avg_psnr', avg_psnr, step)
summary_writer.scalar('train_max_grad_norm', max_grad_norm, step)
summary_writer.scalar('train_avg_grad_norm', avg_grad_norm, step)
summary_writer.scalar('train_max_clipped_grad_norm',
max_clipped_grad_norm, step)
summary_writer.scalar('train_max_grad_max', max_grad_max, step)
summary_writer.scalar('learning_rate', lr, step)
steps_per_sec = config.print_every / (time.time() - t_loop_start)
reset_timer = True
rays_per_sec = config.batch_size * steps_per_sec
summary_writer.scalar('train_steps_per_sec', steps_per_sec, step)
summary_writer.scalar('train_rays_per_sec', rays_per_sec, step)
precision = int(np.ceil(np.log10(config.max_steps))) + 1
print(('{:' + '{:d}'.format(precision) + 'd}').format(step) +
f'/{config.max_steps:d}: ' + f'i_loss={stats.loss[0]:0.4f}, ' +
f'avg_loss={avg_loss:0.4f}, ' +
f'weight_l2={stats.weight_l2[0]:0.2e}, ' + f'lr={lr:0.2e}, ' +
f'{rays_per_sec:0.0f} rays/sec')
if step % config.save_every == 0:
state_to_save = jax.device_get(jax.tree_map(lambda x: x[0], state))
checkpoints.save_checkpoint(
FLAGS.train_dir, state_to_save, int(step), keep=100)
Test-set evaluation
# Test-set evaluation.
if FLAGS.render_every > 0 and step % FLAGS.render_every == 0:
# 我们有意重用优化步骤中的同一个随机数生成器,以便可视化与训练中发生的情况相匹配。.
t_eval_start = time.time()
eval_variables = jax.device_get(jax.tree_map(lambda x: x[0],
state)).optimizer.target
test_case = next(test_dataset)
pred_color, pred_distance, pred_acc = models.render_image(
functools.partial(render_eval_pfn, eval_variables),
test_case['rays'],
keys[0],
chunk=FLAGS.chunk)
vis_suite = vis.visualize_suite(pred_distance, pred_acc)
# Log eval summaries on host 0.
if jax.host_id() == 0:
psnr = math.mse_to_psnr(((pred_color - test_case['pixels'])**2).mean())
ssim = ssim_fn(pred_color, test_case['pixels'])
eval_time = time.time() - t_eval_start
num_rays = jnp.prod(jnp.array(test_case['rays'].directions.shape[:-1]))
rays_per_sec = num_rays / eval_time
summary_writer.scalar('test_rays_per_sec', rays_per_sec, step)
print(f'Eval {step}: {eval_time:0.3f}s., {rays_per_sec:0.0f} rays/sec')
summary_writer.scalar('test_psnr', psnr, step)
summary_writer.scalar('test_ssim', ssim, step)
summary_writer.image('test_pred_color', pred_color, step)
for k, v in vis_suite.items():
summary_writer.image('test_pred_' + k, v, step)
summary_writer.image('test_pred_acc', pred_acc, step)
summary_writer.image('test_target', test_case['pixels'], step)
存储最终的训练模型
if config.max_steps % config.save_every != 0:
state = jax.device_get(jax.tree_map(lambda x: x[0], state))
checkpoints.save_checkpoint(
FLAGS.train_dir, state, int(config.max_steps), keep=100)
def train_step
一个优化步骤
def train_step(model, config, rng, state, batch, lr):
"""One optimization step.
Args:
model: The linen model.
config: The configuration.
rng: jnp.ndarray, random number generator.
state: utils.TrainState, state of the model/optimizer.
batch: dict, a mini-batch of data for training.
lr: float, real-time learning rate.
Returns:
new_state: utils.TrainState, new training state.
stats: list. [(loss, psnr), (loss_coarse, psnr_coarse)].
rng: jnp.ndarray, updated random number generator.
"""
rng, key = random.split(rng)
def loss_fn(variables):
def tree_sum_fn(fn):
return jax.tree_util.tree_reduce(
lambda x, y: x + fn(y), variables, initializer=0)
weight_l2 = config.weight_decay_mult * (
tree_sum_fn(lambda z: jnp.sum(z**2)) /
tree_sum_fn(lambda z: jnp.prod(jnp.array(z.shape))))
ret = model.apply(
variables,
key,
batch['rays'],
randomized=config.randomized,
white_bkgd=config.white_bkgd)
mask = batch['rays'].lossmult
if config.disable_multiscale_loss:
mask = jnp.ones_like(mask)
losses = []
for (rgb, _, _) in ret:
losses.append(
(mask * (rgb - batch['pixels'][..., :3])**2).sum() / mask.sum())
losses = jnp.array(losses)
loss = (
config.coarse_loss_mult * jnp.sum(losses[:-1]) + losses[-1] + weight_l2)
stats = utils.Stats(
loss=loss,
losses=losses,
weight_l2=weight_l2,
psnr=0.0,
psnrs=0.0,
grad_norm=0.0,
grad_abs_max=0.0,
grad_norm_clipped=0.0,
)
return loss, stats
(_, stats), grad = (
jax.value_and_grad(loss_fn, has_aux=True)(state.optimizer.target))
grad = jax.lax.pmean(grad, axis_name='batch')
stats = jax.lax.pmean(stats, axis_name='batch')
def tree_norm(tree):
return jnp.sqrt(
jax.tree_util.tree_reduce(
lambda x, y: x + jnp.sum(y**2), tree, initializer=0))
if config.grad_max_val > 0:
clip_fn = lambda z: jnp.clip(z, -config.grad_max_val, config.grad_max_val)
grad = jax.tree_util.tree_map(clip_fn, grad)
grad_abs_max = jax.tree_util.tree_reduce(
lambda x, y: jnp.maximum(x, jnp.max(jnp.abs(y))), grad, initializer=0)
grad_norm = tree_norm(grad)
if config.grad_max_norm > 0:
mult = jnp.minimum(1, config.grad_max_norm / (1e-7 + grad_norm))
grad = jax.tree_util.tree_map(lambda z: mult * z, grad)
grad_norm_clipped = tree_norm(grad)
new_optimizer = state.optimizer.apply_gradient(grad, learning_rate=lr)
new_state = state.replace(optimizer=new_optimizer)
psnrs = math.mse_to_psnr(stats.losses)
stats = utils.Stats(
loss=stats.loss,
losses=stats.losses,
weight_l2=stats.weight_l2,
psnr=psnrs[-1],
psnrs=psnrs,
grad_norm=grad_norm,
grad_abs_max=grad_abs_max,
grad_norm_clipped=grad_norm_clipped,
)
return new_state, stats, rng
class MipNerfModel
@gin.configurable
class MipNerfModel(nn.Module):
"""Nerf NN Model with both coarse and fine MLPs."""
num_samples: int = 128 # The number of samples per level.
num_levels: int = 2 # The number of sampling levels.
resample_padding: float = 0.01 # Dirichlet/alpha "padding" on the histogram.
stop_level_grad: bool = True # If True, don't backprop across levels')
use_viewdirs: bool = True # If True, use view directions as a condition.
lindisp: bool = False # If True, sample linearly in disparity视差, not in depth.
ray_shape: str = 'cone' # The shape of cast rays ('cone' or 'cylinder'圆柱体).
min_deg_point: int = 0 # Min degree of positional encoding for 3D points.
max_deg_point: int = 16 # Max degree of positional encoding for 3D points.
deg_view: int = 4 # Degree of positional encoding for viewdirs.
density_activation: Callable[..., Any] = nn.softplus # Density activation.
density_noise: float = 0. # Standard deviation of noise added to raw density.
density_bias: float = -1. # The shift added to raw densities pre-activation.
rgb_activation: Callable[..., Any] = nn.sigmoid # The RGB activation.
rgb_padding: float = 0.001 # Padding added to the RGB outputs.
disable_integration: bool = False # If True, use PE instead of IPE.
self值
MipNerfModel(
# attributes
num_samples = 128
num_levels = 2
resample_padding = 0.01
stop_level_grad = True
use_viewdirs = True
lindisp = False
ray_shape = 'cone'
min_deg_point = 0
max_deg_point = 16
deg_view = 4
density_activation = softplus
density_noise = 0.0
density_bias = -1.0
rgb_activation = sigmoid
rgb_padding = 0.001
disable_integration = False
# children
MLP_0 = MLP(
# attributes
net_depth = 8
net_width = 256
net_depth_condition = 1
net_width_condition = 128
net_activation = relu
skip_layer = 4
num_rgb_channels = 3
num_density_channels = 1
)
)
def call
@nn.compact
def __call__(self, rng, rays, randomized, white_bkgd):
"""The mip-NeRF Model.
Args:
rng: jnp.ndarray, random number generator.
rays: util.Rays, a namedtuple命名元组 of ray origins, directions, and viewdirs.
randomized: bool, 使用随机分层抽样.
white_bkgd: bool, if True, use white as the background (black o.w.).
Returns:
ret: list, [*(rgb, distance, acc)]
"""
# Construct the MLP.
mlp = MLP()
ret = []
for i_level in range(self.num_levels):
key, rng = random.split(rng)
分层采样策略
if i_level == 0:
# Stratified sampling along rays
t_vals, samples = mip.sample_along_rays(
key,
rays.origins,
rays.directions,
rays.radii,
self.num_samples,
rays.near,
rays.far,
randomized,
self.lindisp,
self.ray_shape,
)
else:
t_vals, samples = mip.resample_along_rays(
key,
rays.origins,
rays.directions,
rays.radii,
t_vals,
weights,
randomized,
self.ray_shape,
self.stop_level_grad,
resample_padding=self.resample_padding,
)
def sample_along_rays
沿射线分层取样
def sample_along_rays(key, origins, directions, radii, num_samples, near, far,
randomized, lindisp, ray_shape):
"""Stratified sampling along the rays.
Args:
key: jnp.ndarray, random generator key.
origins: jnp.ndarray(float32), [batch_size, 3], ray origins.
directions: jnp.ndarray(float32), [batch_size, 3], ray directions.
radii: jnp.ndarray(float32), [batch_size, 3], ray radii.
num_samples: int.
near: jnp.ndarray, [batch_size, 1], near clip.
far: jnp.ndarray, [batch_size, 1], far clip.
randomized: bool, 使用**随机**分层抽样.
lindisp: bool, 在视差而不是深度上线性采样.
ray_shape: string, 假设光线为哪种形状.
Returns:
t_vals: jnp.ndarray, [batch_size, num_samples], sampled z values.
means: jnp.ndarray, [batch_size, num_samples, 3], sampled means.
covs: jnp.ndarray, [batch_size, num_samples, 3, 3], sampled covariances协方差.
"""
batch_size = origins.shape[0]
t_vals = jnp.linspace(0., 1., num_samples + 1)
if lindisp:
t_vals = 1. / (1. / near * (1. - t_vals) + 1. / far * t_vals)
else:
t_vals = near * (1. - t_vals) + far * t_vals
if randomized:
mids = 0.5 * (t_vals[..., 1:] + t_vals[..., :-1])
upper = jnp.concatenate([mids, t_vals[..., -1:]], -1)
lower = jnp.concatenate([t_vals[..., :1], mids], -1)
t_rand = random.uniform(key, [batch_size, num_samples + 1])
t_vals = lower + (upper - lower) * t_rand
else:
# Broadcast t_vals to make the returned shape consistent.广播t _ vals使返回的形状一致
t_vals = jnp.broadcast_to(t_vals, [batch_size, num_samples + 1])
means, covs = cast_rays(t_vals, origins, directions, radii, ray_shape)
return t_vals, (means, covs)
投射光线(圆锥形或圆柱形)并特征化其截面,求均值和协方差
def cast_rays(t_vals, origins, directions, radii, ray_shape, diag=True):
"""Cast rays (cone- or cylinder-shaped) and featurize sections of it.
Args:
t_vals: float array, the "fencepost" distances along the ray.沿着射线的“fencepost”距离。
origins: float array, the ray origin coordinates.
directions: float array, the ray direction vectors.
radii: float array, the radii (base radii for cones) of the rays.光线的半径(圆锥的底半径)。
ray_shape: string, the shape of the ray, must be 'cone' or 'cylinder'.
diag: boolean, whether or not the covariance matrices should be diagonal.协方差矩阵是否应该是对角的。
Returns:
a tuple of arrays of means and covariances.
"""
t0 = t_vals[..., :-1]
t1 = t_vals[..., 1:]
if ray_shape == 'cone':
gaussian_fn = conical_frustum_to_gaussian
elif ray_shape == 'cylinder':
gaussian_fn = cylinder_to_gaussian
else:
assert False
means, covs = gaussian_fn(directions, t0, t1, radii, diag)
means = means + origins[..., None, :]
return means, covs
t_vals
[[2. 2.03125 2.0625 ... 5.9375 5.96875 6. ]
[2. 2.03125 2.0625 ... 5.9375 5.96875 6. ]
[2. 2.03125 2.0625 ... 5.9375 5.96875 6. ]
...
[2. 2.03125 2.0625 ... 5.9375 5.96875 6. ]
[2. 2.03125 2.0625 ... 5.9375 5.96875 6. ]
[2. 2.03125 2.0625 ... 5.9375 5.96875 6. ]]
位置编码 PE/IPE
self.disable_integration If True, use PE instead of IPE.
if self.disable_integration:
samples = (samples[0], jnp.zeros_like(samples[1]))
samples_enc = mip.integrated_pos_enc(
samples,
self.min_deg_point,
self.max_deg_point,
)
def integrated_pos_enc
用2^[min_deg:max_deg-1缩放的正弦曲线编码“x”
def integrated_pos_enc(x_coord, min_deg, max_deg, diag=True):
"""Encode `x` with sinusoids scaled by 2^[min_deg:max_deg-1].
Args:
x_coord: a tuple containing: x, jnp.ndarray, variables to be encoded. Should
be in [-pi, pi]. x_cov, jnp.ndarray, covariance matrices for `x`.
min_deg: int, the min degree of the encoding.
max_deg: int, the max degree of the encoding.
diag: bool, if true, expects input covariances to be diagonal (full
otherwise).
Returns:
encoded: jnp.ndarray, encoded variables.
"""
if diag:
x, x_cov_diag = x_coord
scales = jnp.array([2**i for i in range(min_deg, max_deg)])
shape = list(x.shape[:-1]) + [-1]
y = jnp.reshape(x[..., None, :] * scales[:, None], shape)
y_var = jnp.reshape(x_cov_diag[..., None, :] * scales[:, None]**2, shape)
else:
x, x_cov = x_coord
num_dims = x.shape[-1]
basis = jnp.concatenate(
[2**i * jnp.eye(num_dims) for i in range(min_deg, max_deg)], 1)
y = math.matmul(x, basis)
# Get the diagonal of a covariance matrix (ie, variance). This is equivalent
# to jax.vmap(jnp.diag)((basis.T @ covs) @ basis).
y_var = jnp.sum((math.matmul(x_cov, basis)) * basis, -2)
return expected_sin(
jnp.concatenate([y, y + 0.5 * jnp.pi], axis=-1),
jnp.concatenate([y_var] * 2, axis=-1))[0]
def expected_sin(x, x_var):
"""Estimates mean and variance of sin(z), z ~ N(x, var)."""
# 当方差很大时,将sin向零收缩.
y = jnp.exp(-0.5 * x_var) * math.safe_sin(x)
y_var = jnp.maximum(
0, 0.5 * (1 - jnp.exp(-2 * x_var) * math.safe_cos(2 * x)) - y**2)
return y, y_var
点属性预测 raw_rgb, raw_density
raw_rgb, raw_density = mlp(samples_enc, viewdirs_enc)
raw_rgb, raw_density = mlp(samples_enc)
# Point attribute predictions
if self.use_viewdirs:
viewdirs_enc = mip.pos_enc(
rays.viewdirs,
min_deg=0,
max_deg=self.deg_view,
append_identity=True,
)
raw_rgb, raw_density = mlp(samples_enc, viewdirs_enc)
else:
raw_rgb, raw_density = mlp(samples_enc)
def pos_enc(x, min_deg, max_deg, append_identity=True):
"""The positional encoding used by the original NeRF paper."""
scales = jnp.array([2**i for i in range(min_deg, max_deg)])
xb = jnp.reshape((x[..., None, :] * scales[:, None]),
list(x.shape[:-1]) + [-1])
four_feat = jnp.sin(jnp.concatenate([xb, xb + 0.5 * jnp.pi], axis=-1))
if append_identity:
return jnp.concatenate([x] + [four_feat], axis=-1)
else:
return four_feat
体积渲染
如果需要,添加噪音以调整密度预测。
# Add noise to regularize the density predictions if needed.
if randomized and (self.density_noise > 0):
key, rng = random.split(rng)
raw_density += self.density_noise * random.normal(
key, raw_density.shape, dtype=raw_density.dtype)
# Volumetric rendering.
rgb = self.rgb_activation(raw_rgb)
rgb = rgb * (1 + 2 * self.rgb_padding) - self.rgb_padding
density = self.density_activation(raw_density + self.density_bias)
comp_rgb, distance, acc, weights = mip.volumetric_rendering(
rgb,
density,
t_vals,
rays.directions,
white_bkgd=white_bkgd,
)
ret.append((comp_rgb, distance, acc))
return ret
ret
两个level 三个属性值(comp_rgb, distance, acc)

def volumetric_rendering
def volumetric_rendering(rgb, density, t_vals, dirs, white_bkgd):
"""Volumetric Rendering Function.
Args:
rgb: jnp.ndarray(float32), color, [batch_size, num_samples, 3]
density: jnp.ndarray(float32), density, [batch_size, num_samples, 1].
t_vals: jnp.ndarray(float32), [batch_size, num_samples].
dirs: jnp.ndarray(float32), [batch_size, 3].
white_bkgd: bool.
Returns:
comp_rgb: jnp.ndarray(float32), [batch_size, 3].
disp: jnp.ndarray(float32), [batch_size].
acc: jnp.ndarray(float32), [batch_size].
weights: jnp.ndarray(float32), [batch_size, num_samples]
"""
t_mids = 0.5 * (t_vals[..., :-1] + t_vals[..., 1:])
t_dists = t_vals[..., 1:] - t_vals[..., :-1]
delta = t_dists * jnp.linalg.norm(dirs[..., None, :], axis=-1)
# Note that we're quietly turning density from [..., 0] to [...].
density_delta = density[..., 0] * delta
alpha = 1 - jnp.exp(-density_delta)
trans = jnp.exp(-jnp.concatenate([
jnp.zeros_like(density_delta[..., :1]),
jnp.cumsum(density_delta[..., :-1], axis=-1)
],
axis=-1))
weights = alpha * trans
comp_rgb = (weights[..., None] * rgb).sum(axis=-2)
acc = weights.sum(axis=-1)
distance = (weights * t_mids).sum(axis=-1) / acc
distance = jnp.clip(
jnp.nan_to_num(distance, jnp.inf), t_vals[:, 0], t_vals[:, -1])
if white_bkgd:
comp_rgb = comp_rgb + (1. - acc[..., None])
return comp_rgb, distance, acc, weights
class MLP
@gin.configurable
class MLP(nn.Module):
"""A simple MLP."""
net_depth: int = 8 # The depth of the first part of MLP.
net_width: int = 256 # The width of the first part of MLP.
net_depth_condition: int = 1 # The depth of the second part of MLP.
net_width_condition: int = 128 # The width of the second part of MLP.
net_activation: Callable[..., Any] = nn.relu # The activation function.
skip_layer: int = 4 # Add a skip connection to the output of every N layers.向每N层的输出添加一个跳过连接
num_rgb_channels: int = 3 # The number of RGB channels.
num_density_channels: int = 1 # The number of density channels.
def call
@nn.compact
def __call__(self, x, condition=None):
"""Evaluate the MLP.
Args:
x: jnp.ndarray(float32), [batch, num_samples, feature], points.
condition: jnp.ndarray(float32), [batch, feature], if not None, this
variable will be part of the input to the second part of the MLP
concatenated with the output vector of the first part of the MLP. If
None, only the first part of the MLP will be used with input x. In the
original paper, this variable is the view direction.
Returns:
raw_rgb: jnp.ndarray(float32), with a shape of
[batch, num_samples, num_rgb_channels].
raw_density: jnp.ndarray(float32), with a shape of
[batch, num_samples, num_density_channels].
"""
feature_dim = x.shape[-1]
num_samples = x.shape[1]
x = x.reshape([-1, feature_dim])
dense_layer = functools.partial(
nn.Dense, kernel_init=jax.nn.initializers.glorot_uniform())
inputs = x
for i in range(self.net_depth):
x = dense_layer(self.net_width)(x)
x = self.net_activation(x)
if i % self.skip_layer == 0 and i > 0:
x = jnp.concatenate([x, inputs], axis=-1)
raw_density = dense_layer(self.num_density_channels)(x).reshape(
[-1, num_samples, self.num_density_channels])
if condition is not None:
# Output of the first part of MLP.
bottleneck = dense_layer(self.net_width)(x)
# Broadcast condition from [batch, feature] to
# [batch, num_samples, feature] since all the samples along the same ray
# have the same viewdir.
condition = jnp.tile(condition[:, None, :], (1, num_samples, 1))
# Collapse the [batch, num_samples, feature] tensor to
# [batch * num_samples, feature] so that it can be fed into nn.Dense.
condition = condition.reshape([-1, condition.shape[-1]])
x = jnp.concatenate([bottleneck, condition], axis=-1)
# Here use 1 extra layer to align with the original nerf model.
for i in range(self.net_depth_condition):
x = dense_layer(self.net_width_condition)(x)
x = self.net_activation(x)
raw_rgb = dense_layer(self.num_rgb_channels)(x).reshape(
[-1, num_samples, self.num_rgb_channels])
return raw_rgb, raw_density
jax
pmap
train_pstep = jax.pmap(
functools.partial(train_step, model, config),
axis_name='batch',
in_axes=(0, 0, 0, None),
donate_argnums=(2,))
支持集体行动的并行映射
def pmap(
fun: F,
axis_name: Optional[AxisName] = None,
*,
in_axes=0,
out_axes=0,
static_broadcasted_argnums: Union[int, Iterable[int]] = (),
devices: Optional[Sequence[xc.Device]] = None,
backend: Optional[str] = None,
axis_size: Optional[int] = None,
donate_argnums: Union[int, Iterable[int]] = (),
global_arg_shapes: Optional[Tuple[Tuple[int, ...], ...]] = None,
) -> F:
"""Parallel map with support for collective operations.
The purpose of :py:func:`pmap` 是表示单程序多数据 (SPMD)程序. Applying :py:func:`pmap` to a function
将编译 函数(类似于:py:func:`jit ` ),然后并行执行它 在XLA设备上,例如多个GPU或多个TPU核心.
语义上它与:py:func:`vmap '相当,因为两种转换都映射一个函数 阵列轴上,
但是 :py:func:`vmap '通过将 将轴向下映射到原始操作,
:py:func:`pmap '改为复制 函数,并在其自己的XLA设备上并行执行每个复制副本。
映射的轴大小必须小于或等于本地XLA的数量 可用设备, 如:py:func:` jax . local _ device _ count()`(除非 指定了“设备”,见下文).
对于嵌套的:py:func:`pmap '调用,映射轴大小的乘积必须小于或等于XLA设备的数量。
.. note::
:py:func:`pmap` compiles ``fun``, 因此,虽然它可以与:py:func:`jit '结合使用,但通常是不必要的。
**Multi-process platforms:** On multi-process platforms such as TPU pods,
:py:func:`pmap` is designed to be used in SPMD Python programs, where every
process is running the same Python code such that all processes run the same
pmapped function in the same order. Each process should still call the pmapped
function with mapped axis size equal to the number of *local* devices (unless
``devices`` is specified, see below), and an array of the same leading axis
size will be returned as usual. However, any collective operations in ``fun``
will be computed over *all* participating devices, including those on other
processes, via device-to-device communication. Conceptually, this can be
thought of as running a pmap over a single array sharded across processes,
where each process "sees" only its local shard of the input and output. The
SPMD model requires that the same multi-process pmaps must be run in the same
order on all devices, but they can be interspersed with arbitrary operations
running in a single process.
Args:
fun: Function to be mapped over argument axes. Its arguments and return
value should be arrays, scalars, or (nested) standard Python containers
(tuple/list/dict) thereof. Positional arguments indicated by
``static_broadcasted_argnums`` can be anything at all, provided they are
hashable and have an equality operation defined.
axis_name: Optional, a hashable Python object used to identify the mapped
axis so that parallel collectives can be applied.
in_axes: A non-negative integer, None, or nested Python container thereof
that specifies which axes of positional arguments to map over. Arguments
passed as keywords are always mapped over their leading axis (i.e. axis
index 0). See :py:func:`vmap` for details.
out_axes: A non-negative integer, None, or nested Python container thereof
indicating where the mapped axis should appear in the output. All outputs
with a mapped axis must have a non-None ``out_axes`` specification
(see :py:func:`vmap`).
static_broadcasted_argnums: An int or collection of ints specifying which
positional arguments to treat as static (compile-time constant).
Operations that only depend on static arguments will be constant-folded.
Calling the pmapped function with different values for these constants
will trigger recompilation. If the pmapped function is called with fewer
positional arguments than indicated by ``static_argnums`` then an error is
raised. Each of the static arguments will be broadcasted to all devices.
Arguments that are not arrays or containers thereof must be marked as
static. Defaults to ().
devices: This is an experimental feature and the API is likely to change.
Optional, a sequence of Devices to map over. (Available devices can be
retrieved via jax.devices()). Must be given identically for each process
in multi-process settings (and will therefore include devices across
processes). If specified, the size of the mapped axis must be equal to
the number of devices in the sequence local to the given process. Nested
:py:func:`pmap` s with ``devices`` specified in either the inner or outer
:py:func:`pmap` are not yet supported.
backend: This is an experimental feature and the API is likely to change.
Optional, a string representing the XLA backend. 'cpu', 'gpu', or 'tpu'.
axis_size: Optional; the size of the mapped axis.
donate_argnums: Specify which arguments are "donated" to the computation.
It is safe to donate arguments if you no longer need them once the
computation has finished. In some cases XLA can make use of donated
buffers to reduce the amount of memory needed to perform a computation,
for example recycling one of your input buffers to store a result. You
should not reuse buffers that you donate to a computation, JAX will raise
an error if you try to.
global_arg_shapes: Optional, must be set when using pmap(sharded_jit) and
the partitioned values span multiple processes. The global cross-process
per-replica shape of each argument, i.e. does not include the leading
pmapped dimension. Can be None for replicated arguments. This API is
likely to change in the future.
Returns:
A parallelized version of ``fun`` with arguments that correspond to those of
``fun`` but with extra array axes at positions indicated by ``in_axes`` and
with output that has an additional leading array axis (with the same size).
For example, assuming 8 XLA devices are available, :py:func:`pmap` can be used
as a map along a leading array axis:
>>> import jax.numpy as jnp
>>>
>>> out = pmap(lambda x: x ** 2)(jnp.arange(8)) # doctest: +SKIP
>>> print(out) # doctest: +SKIP
[0, 1, 4, 9, 16, 25, 36, 49]
When the leading dimension is smaller than the number of available devices JAX
will simply run on a subset of devices:
>>> x = jnp.arange(3 * 2 * 2.).reshape((3, 2, 2))
>>> y = jnp.arange(3 * 2 * 2.).reshape((3, 2, 2)) ** 2
>>> out = pmap(jnp.dot)(x, y) # doctest: +SKIP
>>> print(out) # doctest: +SKIP
[[[ 4. 9.]
[ 12. 29.]]
[[ 244. 345.]
[ 348. 493.]]
[[ 1412. 1737.]
[ 1740. 2141.]]]
If your leading dimension is larger than the number of available devices you
will get an error:
>>> pmap(lambda x: x ** 2)(jnp.arange(9)) # doctest: +SKIP
ValueError: ... requires 9 replicas, but only 8 XLA devices are available
As with :py:func:`vmap`, using ``None`` in ``in_axes`` indicates that an
argument doesn't have an extra axis and should be broadcasted, rather than
mapped, across the replicas:
>>> x, y = jnp.arange(2.), 4.
>>> out = pmap(lambda x, y: (x + y, y * 2.), in_axes=(0, None))(x, y) # doctest: +SKIP
>>> print(out) # doctest: +SKIP
([4., 5.], [8., 8.])
Note that :py:func:`pmap` always returns values mapped over their leading axis,
equivalent to using ``out_axes=0`` in :py:func:`vmap`.
In addition to expressing pure maps, :py:func:`pmap` can also be used to express
parallel single-program multiple-data (SPMD) programs that communicate via
collective operations. For example:
>>> f = lambda x: x / jax.lax.psum(x, axis_name='i')
>>> out = pmap(f, axis_name='i')(jnp.arange(4.)) # doctest: +SKIP
>>> print(out) # doctest: +SKIP
[ 0. 0.16666667 0.33333334 0.5 ]
>>> print(out.sum()) # doctest: +SKIP
1.0
In this example, ``axis_name`` is a string, but it can be any Python object
with ``__hash__`` and ``__eq__`` defined.
The argument ``axis_name`` to :py:func:`pmap` names the mapped axis so that
collective operations, like :func:`jax.lax.psum`, can refer to it. Axis names
are important particularly in the case of nested :py:func:`pmap` functions,
where collective operations can operate over distinct axes:
>>> from functools import partial
>>> import jax
>>>
>>> @partial(pmap, axis_name='rows')
... @partial(pmap, axis_name='cols')
... def normalize(x):
... row_normed = x / jax.lax.psum(x, 'rows')
... col_normed = x / jax.lax.psum(x, 'cols')
... doubly_normed = x / jax.lax.psum(x, ('rows', 'cols'))
... return row_normed, col_normed, doubly_normed
>>>
>>> x = jnp.arange(8.).reshape((4, 2))
>>> row_normed, col_normed, doubly_normed = normalize(x) # doctest: +SKIP
>>> print(row_normed.sum(0)) # doctest: +SKIP
[ 1. 1.]
>>> print(col_normed.sum(1)) # doctest: +SKIP
[ 1. 1. 1. 1.]
>>> print(doubly_normed.sum((0, 1))) # doctest: +SKIP
1.0
On multi-process platforms, collective operations operate over all devices,
including those on other processes. For example, assuming the following code
runs on two processes with 4 XLA devices each:
>>> f = lambda x: x + jax.lax.psum(x, axis_name='i')
>>> data = jnp.arange(4) if jax.process_index() == 0 else jnp.arange(4, 8)
>>> out = pmap(f, axis_name='i')(data) # doctest: +SKIP
>>> print(out) # doctest: +SKIP
[28 29 30 31] # on process 0
[32 33 34 35] # on process 1
Each process passes in a different length-4 array, corresponding to its 4
local devices, and the psum operates over all 8 values. Conceptually, the two
length-4 arrays can be thought of as a sharded length-8 array (in this example
equivalent to jnp.arange(8)) that is mapped over, with the length-8 mapped
axis given name 'i'. The pmap call on each process then returns the
corresponding length-4 output shard.
The ``devices`` argument can be used to specify exactly which devices are used
to run the parallel computation. For example, again assuming a single process
with 8 devices, the following code defines two parallel computations, one
which runs on the first six devices and one on the remaining two:
>>> from functools import partial
>>> @partial(pmap, axis_name='i', devices=jax.devices()[:6])
... def f1(x):
... return x / jax.lax.psum(x, axis_name='i')
>>>
>>> @partial(pmap, axis_name='i', devices=jax.devices()[-2:])
... def f2(x):
... return jax.lax.psum(x ** 2, axis_name='i')
>>>
>>> print(f1(jnp.arange(6.))) # doctest: +SKIP
[0. 0.06666667 0.13333333 0.2 0.26666667 0.33333333]
>>> print(f2(jnp.array([2., 3.]))) # doctest: +SKIP
[ 13. 13.]
"""
# axis_size is an optional integer representing the global axis size. The
# aggregate size (across all processes) size of the mapped axis must match the
# given value.