医学图像分割之肝脏分割(2D)
文章目录
- 肝脏分割数据集
- 一、预处理
- 二、图像增强
- 三、训练
- 四、测试
- 总结
肝脏分割数据集
肝脏分割分割主要有三个开源数据集:LiTS17,Sliver07,3DIRCADb
LiTS17包含131个数据,可以用于肝脏及肿瘤的分割
Sliver07包含20个数据,但仅仅包含肝脏,并没有肿瘤的金标准
3DIRCADb包含20个数据,可以用于肝脏及肿瘤的分割
提示:以下是本篇文章正文内容,下面案例可供参考
一、预处理
这三个肝脏分割数据集都是3D数据,所以如过要进行训练必须要对数据进行切片。
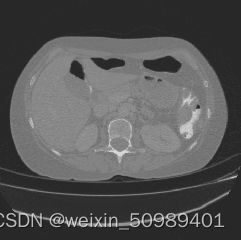
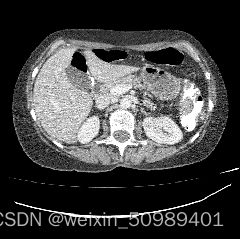
对3D nii文件进行调窗,找到肝脏区域并向外扩张,将数据z轴的spacing调整到1mm。
将3D nii格式的数据切片为2D png格式。
# 将灰度值在阈值之外的截断掉
# upper = 200
# lower = -200
ct_array[ct_array > para.upper] = para.upper
ct_array[ct_array < para.lower] = para.lower
#肝脏分割设置
seg_array[seg_array > 0] = 1
# 找到肝脏区域开始和结束的slice,并各向外扩张
z = np.any(seg_array, axis=(1, 2))
start_slice, end_slice = np.where(z)[0][[0, -1]]
# 对CT数据在横断面上进行降采样(下采样),并进行重采样,将所有数据的z轴的spacing调整到1mm
ct_array = ndimage.zoom(ct_array,(ct.GetSpacing()[-1] / para.slice_thickness, para.down_scale, para.down_scale),order=3)
seg_array = ndimage.zoom(seg_array, (ct.GetSpacing()[-1] / para.slice_thickness, para.down_scale, para.down_scale), order=0)
#进行切片操作
for i in range(volume.shape[0]):
slice = volume[i,:,:]
slice_post = WL(slice, WC, WW)
slice_post = slice_post.astype(np.uint8)
# slice_post = cv.equalizeHist(slice_post)
slices_in_order.append(slice_post)原始CT 调窗后
二、图像增强
同时对肝脏的 Original CT,Ground Truth进行图像增强。
# 图像旋转: 按照概率0.8执行,最大左旋角度10,最大右旋角度10
# p.rotate(probability=0.5, max_left_rotation=20, max_right_rotation=20)
# 图像左右互换: 按照概率0.5执行
# p.flip_left_right(probability=0.8)
# # # 图像放大缩小: 按照概率0.8执行,面积为原始图0.85倍
# p.zoom_random(probability=1, percentage_area=0.8)三、训练
1.模型搭建
import torch.nn as nn
import torch
from torch import autograd
class DoubleConv(nn.Module):
def __init__(self, in_ch, out_ch):
super(DoubleConv, self).__init__()
self.conv = nn.Sequential(
nn.Conv2d(in_ch, out_ch, 3, padding=1),
nn.BatchNorm2d(out_ch),
nn.ReLU(inplace=True),
nn.Conv2d(out_ch, out_ch, 3, padding=1),
nn.BatchNorm2d(out_ch),
nn.ReLU(inplace=True)
)
def forward(self, input):
return self.conv(input)
class UNet(nn.Module):
def __init__(self, in_ch, out_ch):
super(UNet, self).__init__()
self.conv1 = DoubleConv(in_ch, 64)
self.pool1 = nn.MaxPool2d(2)
self.conv2 = DoubleConv(64, 128)
self.pool2 = nn.MaxPool2d(2)
self.conv3 = DoubleConv(128, 256)
self.pool3 = nn.MaxPool2d(2)
self.conv4 = DoubleConv(256, 512)
self.pool4 = nn.MaxPool2d(2)
self.conv5 = DoubleConv(512, 1024)
self.up6 = nn.ConvTranspose2d(1024, 512, 2, stride=2)
self.conv6 = DoubleConv(1024, 512)
self.up7 = nn.ConvTranspose2d(512, 256, 2, stride=2)
self.conv7 = DoubleConv(512, 256)
self.up8 = nn.ConvTranspose2d(256, 128, 2, stride=2)
self.conv8 = DoubleConv(256, 128)
self.up9 = nn.ConvTranspose2d(128, 64, 2, stride=2)
self.conv9 = DoubleConv(128, 64)
self.conv10 = nn.Conv2d(64, out_ch, 1)
def forward(self, x):
c1 = self.conv1(x)
p1 = self.pool1(c1)
c2 = self.conv2(p1)
p2 = self.pool2(c2)
c3 = self.conv3(p2)
p3 = self.pool3(c3)
c4 = self.conv4(p3)
p4 = self.pool4(c4)
c5 = self.conv5(p4)
up_6 = self.up6(c5)
merge6 = torch.cat([up_6, c4], dim=1)
c6 = self.conv6(merge6)
up_7 = self.up7(c6)
merge7 = torch.cat([up_7, c3], dim=1)
c7 = self.conv7(merge7)
up_8 = self.up8(c7)
merge8 = torch.cat([up_8, c2], dim=1)
c8 = self.conv8(merge8)
up_9 = self.up9(c8)
merge9 = torch.cat([up_9, c1], dim=1)
c9 = self.conv9(merge9)
c10 = self.conv10(c9)
out = nn.Sigmoid()(c10)
return out2.模型训练
if __name__ == '__main__':
device = torch.device("cuda" if torch.cuda.is_available() else "cpu")
LEARNING_RATE = 1e-3
LR_DECAY_STEP = 2
LR_DECAY_FACTOR = 0.5
WEIGHT_DECAY = 5e-4
BATCH_SIZE = 1
MAX_EPOCHS = 2
model = UNet()
criterion = DiceLoss()
if opt.use_gpu:
criterion = criterion.cuda(opt.device)
lr = opt.lr
optimizer = torch.optim.Adam(model.parameters(), lr=lr, weight_decay=opt.weight_decay)
lr_scheduler = torch.optim.lr_scheduler.ExponentialLR(optimizer, opt.lr_decay)
MODEL = Unet(out_ch=2)
OPTIMIZER = torch.optim.Adam(MODEL.parameters(), lr=LEARNING_RATE, weight_decay=WEIGHT_DECAY)
LR_SCHEDULER = torch.optim.lr_scheduler.StepLR(OPTIMIZER, step_size=LR_DECAY_STEP, gamma=LR_DECAY_FACTOR)
CRITERION = DiceLoss().to(device)四、测试
for i in range(1, 10):
idx_list.append(i)
ct = sitk.ReadImage(data_path + 'volume-' + str(i) + '.nii',sitk.sitkInt16)
seg = sitk.ReadImage(data_path + 'segmentation-' + str(i) + '.nii',sitk.sitkInt16)
predictions_in_order = []
for slice in slices_in_order:
slice = torch.from_numpy(slice).float() / 255.
output = model(slice.unsqueeze(0).unsqueeze(0))
prediction = sm(output)
_, prediction = torch.max(prediction, dim=1) #返回每一行中最大值的那个元素,且返回其索引(返回最大元素在这一行的列索引)
prediction = prediction.squeeze(0).cpu().detach().numpy().astype(np.uint8)
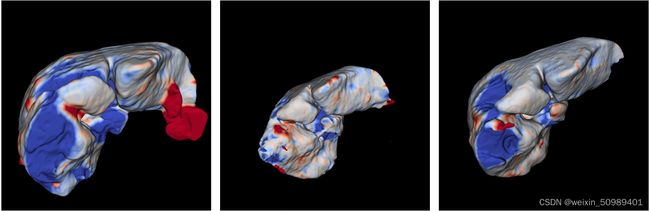
predictions_in_order.append(prediction)测试结果可视化:
总结
第一次写,也是随手一写,后续会更新3D肝脏分割,2.5D肝脏分割,以及肝脏肿瘤分割。