R语言完成t-SNE分析和画图
1.编一个数据
需要一个行为样本,列为变量(例如基因)的矩阵。
dat = matrix(rnorm(6000),ncol = 20)
dat[101:200,] = dat[101:200,]+rnorm(1,10,10)
dat[201:300,] = dat[201:300,]+rnorm(1,15,15)
rownames(dat) = paste0("a",1:300)
colnames(dat) = paste0("g",1:20)
dat[1:4,1:4]
## g1 g2 g3 g4
## a1 -0.149414 1.1634082 -0.6643721 0.49231425
## a2 1.241976 0.2490248 -0.5021437 0.75851991
## a3 1.370120 1.0039089 -1.4565175 -0.60521941
## a4 -1.151987 -0.2600440 -0.3445524 -0.05260084
pheatmap::pheatmap(dat,show_rownames = F,color = colorRampPalette(c("navy", "white", "firebrick3"))(50))
2.完成tsne分析并画图
library(Rtsne)
tsne_out = Rtsne(dat,perplexity = 30)
pdat = data.frame(tsne_out$Y,rep(c("a","b","c"),each = 100))
colnames(pdat) = c("Y1","Y2","group")
head(pdat)
## Y1 Y2 group
## 1 -2.053756 -14.37403 a
## 2 -1.952955 -12.66466 a
## 3 -2.837022 -13.36159 a
## 4 -4.512561 -12.64833 a
## 5 -2.803907 -10.89250 a
## 6 -1.956784 -12.09723 a
画图搞起
library(ggplot2)
ggplot(pdat,aes(Y1,Y2))+
geom_point(aes(Y1,Y2,fill = group),shape = 21,color = "black")+
stat_ellipse(aes(color = group,fill = group),
geom = "polygon",
alpha = 0.3,
linetype = 2)+
theme_classic()+
theme(legend.position = "top")
3.神奇参数perplexity
perplexity的默认值是30。样本数量比较小的时候,会报个错说 Error in .check_tsne_params(nrow(X), dims = dims, perplexity = perplexity, : perplexity is too large for the number of samples
一开始我拿的示例数据20行和40行时,都会报这个错。解决办法很简单,把这个参数调小一些即可。查了一下关于这个参数的说明,有几个结果值得记录:
3.1.什么是perplexity
直译是“困惑”,啊这。。确实困惑。
对它的解释是:
This value effectively controls how many nearest neighbours are taken into account when constructing the embedding in the low-dimensional space.
–出自帮助文档
3.2.合理取值范围
The performance of SNE is fairly robust to changes in the perplexity, and typical values are between 5 and 50.
–出自tsne论文原文
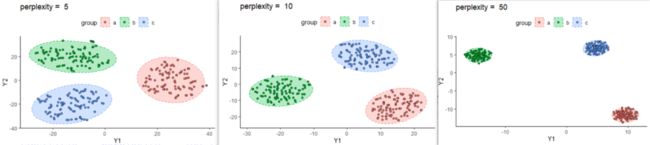
3.3. perplexity变化对结果的影响
对于我们这个示例数据,影响确实不大。5,10,50的结果如下,当perplexity = 100的时候就报错了。
有人对这个参数进行了更加深入的探索,值得一读,见https://distill.pub/2016/misread-tsne/。