VIDHOP, viral host prediction with Deep Learning 论文阅读笔记
VIDHOP, viral host prediction with Deep Learning 论文阅读笔记
github : https://github.com/flomock/vidhop
摘要
Zoonosis即人类和动物都可相互传染致病,如寨卡病毒、埃博拉病毒和新冠病毒等,为了预防全球化带来的病毒传染加快的问题,本文提出一种基于病毒的基因组序列来推测病毒宿主的预测方法(input = 病毒基因碱基序列, Y = 宿主种类),并且定义了一种基于预测宿主数来计算的平均准确度的计算公式,本模型可以用于transfer到其他的病毒上面去,直接分类准确度起伏较大,基于作者定义的准确度比较平稳。
原理
模型总览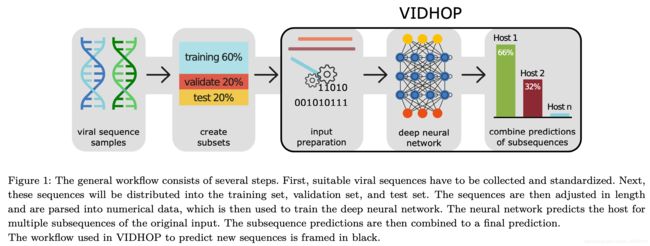
如上图所示,先从国家某些生物网站上下载数据——本文中从欧洲核苷酸档案 (ENA) 数据库 收集了带有宿主标签的甲型流感病毒、狂犬病狂犬病病毒和轮状病毒 A 的所有核苷酸序列,再从德国生物技术信息中心 (NCBI) 提供的分类信息来管理宿主标签——然后将其按照6:2:2来划分成训练集、验证集和测试集,在将数据进行预处理输入模型得到预测结果并且进行分析,下文将对模型每一个步骤进行深入的介绍。
数据预处理
数据预处理的要点在于每一个病毒基因序列的长短都是不一样的,当数据集一旦变大,这个现象带来的影响就被放大。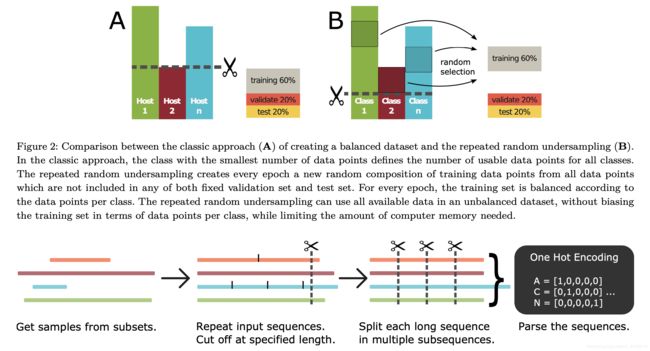
所以就要用到上图所示的方法对数据集进行修剪,修剪的长度等于基因从小到大排序的95%分位的长度,这样就可以很好地把数据统一起来。
但是有个问题是,如果最短基因序列的长度过于短,这样修剪出来的数据放到模型里面训练就会欠拟合,所以本文针对这一问题提出了几种数据扩展方案:
- 简单重复:将原基因本体重复拼接在后面以达到规定长度
- 简单重复并填充占位符:在上一个方法的基础上在任意位置随机插入2-8个占位符
- 随机重复:随机抽取原基因的一部分重复拼接到原基因后面
- 随机重复并填充占位符:同上类比
- 填充占位符:在原基因的末尾全部填充占位符到规定长度
- 裁剪:将全部基因的长度修剪到最短基因的长度
处理好之后的数据集在输入到模型的时候为了加速训练的过程,将其拆分成小片分批依次输入到模型里面去。最后要对多批次的输出结果进行处理得到一个最终的预测结果。对碱基进行one-hot编码,如:A = [1, 0, 0, 0, 0], T = [0, 0, 0, 1, 0], N = [0, 0, 0, 0, 1]。
本文还提出了一种叫做在线学习(Online)的方法,这种方法并不需要在输入模型训练的时候对数据进行预处理,而是在训练的过程中用修改过的数据对模型施加一定的影响,具体操作方法见实际代码,文中并没有加以详细描述。
在训练的时候,可以划分好一定数量的验证集和测试集,基于剩下的数据,每一个训练epoch都抽取一定数量(小于剩下样本的总数)不同的样本进行训练,这叫做随机重复欠采样,可以避免因为某个类别的数据过多引起的训练“注意力偏移”。
DNN模型结构
本文着重提出了两种DNN模型,一种是纯LSTM模型,另一种是CNN+LSTM模型,他们的差别在于前者是由三层LSTM接两层Dense,后者是两层全连接层+两层LSTM+两层Dense,最后一层Dense用于分类。LSTM可以很好的应对复杂的数据集和处理任务,CNN则相对于LSTM来说计算速度是其四倍。对于类别种类很多的模型来说,数据量很大,所以就更加需要之前提到的输入切片。
预测结果处理
由于输入用的是切片输入,一个长的基因被切成好几段长分别输入到模型里面,每一段都会有相应的预测结果,所以要把全部结果整合到一起去才能代表整个一个基因的预测结果,所以本文提出了几种方法:
- 不处理:直接输出每个基因切片的预测结果
- 票选法: 对所有子序列使用多数“投票”来确定预测结果,每个子序列的票就是其所属类别。这种方法得出的结果偏差较大比较“离散”,不具有一般连续性。
- 均值法:将所有子序列的概率按类取平均值,最后得出平均值大的那一类作为最终结果。
- 标准差:在上以种方法的基础上,用其标准差对每个子序列进行加权。 具有更多不同预测类别的子序列获得更高的权重。
最后的结果要通过以上方法处理之后才能得到。
实验结果
概况
实验用了两种模型针对三个不同的数据集分别进行,最多训练了1500个epochs,当acc在300个epochs还没上升的情况下,模型停止训练。对于轮状病毒A数据集来说,数据集总共有40000个病毒基因序列,对应6个宿主类别。因为这几个宿主类别之间关联性比较强,实验测得6个不同的宿主导致预期的随机准确度约为 16.67%,导致两个模型直接测得的预测准确率都比较高,分别是85.28%和82.88%,而在甲流病毒数据集中宿主类别有36个,36个不同的宿主导致预期的随机准确度只有2.78%,所以直接测得的准确率只有50%左右。所以可以得出一个结论:宿主数量越多类型差别越大,随机准确率越低。
实验结果如下表所示
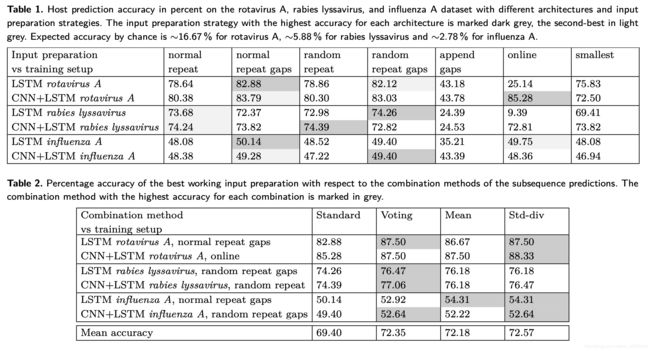
不难看出,在诸多预处理数据扩展方案中,最有效的是简单重复并填充占位符和随机重复并填充占位符,其他的方案效果不佳。
为了通过考虑类的数量获得更好的可比性,所以本文引入了一个新的准确率计算公式:
average accuracy = 2 ⋅ accuracy + ∣ classes ∣ − 2 ∣ classes ∣ \text{average accuracy} =\frac{2 \cdot \text { accuracy }+\mid \text { classes } \mid-2}{\mid \text { classes } \mid} average accuracy=∣ classes ∣2⋅ accuracy +∣ classes ∣−2
通过这个公式就可以修正分类准确率,如下表示
与其他模型的对比请看原文
总结
本文提出了一种使用病毒碱基序列编码成数据用于训练的模型来预测分类宿主,并用了几个不同的数据集进行实验,结果表明当分类的类别增大的时候,想要准确预测宿主类别就变得比较困难,本文中使用的模型结构比较简单,并没有用上现在最新的模型。本文的主要贡献其一也就是本文花大量篇幅来讲解的数据处理部分,在对基因序列进行修剪之后怎么样对它们进行处理来让他们变成可用的数据;其二就是为我们引出了一个新的方向,关于使用NLP的方法来处理基因序列,从而可以训练模型来预测给定基因对应的宿主类型,在现在新冠流行的大环境下是一个比较有意义的事情。
2021.07.12更新
实验流程及对应代码解读
代码结构
vidhop
|-- DataParsing
| `-- DataParsing_main.py
|-- cli.py
|-- training
| |-- DataGenerator.py
| |-- make_dataset_out
| | |-- X_test.csv
| | |-- X_train.csv
| | |-- X_val.csv
| | |-- Y_test.csv
| | |-- Y_train.csv
| | `-- Y_val.csv
| |-- make_datasets.py
| `-- train_new_model.py
|-- vidhop_main.py
`-- weights
|-- influ_weights.best.acc.normal_repeat_spacer_run2.hdf5
|-- rabies_weights.best.acc.random_repeat_run2_design_7.hdf5
`-- rota_weights.best.acc.online_design_7.hdf5
整个项目的结构如上图所示,其中:
- DataParsing 部分用于预处理数据
- DataGenerator.py 用于提供训练模型的数据
- make_dataset.py 用于从数据集中抽取并划分train、val和test的数据,并写入csv文件,生成用于预处理的数据
- train_new_model.py 用于训练模型,里面包含了训练模型、处理数据等全部过程
- vidhop_main.py 用于加载模型来进行测试
- weights 文件夹内保存的是这个模型的预训练权重
数据预处理——DataParsing.py
class CircularList(list): # 获取list
def __getitem__(self, x):
if isinstance(x, slice):
return [self[x] for x in self._rangeify(x)]
index = operator.index(x)
try:
return super().__getitem__(index % len(self))
except ZeroDivisionError:
raise IndexError('list index out of range')
def _rangeify(self, slice):
start, stop, step = slice.start, slice.stop, slice.step
if start is None:
start = 0
if stop is None:
stop = len(self)
if step is None:
step = 1
return range(start, stop, step)
获取待处理的list
def encode_string(maxLen=None, x=[], y=[], y_encoder=None, repeat=True, use_spacer=False, online_Xtrain_set=False,
randomrepeat=False):
"""
One hot encoding for classes
to convert the "old" exported int data via OHE to binary matrix
http://machinelearningmastery.com/multi-class-classification-tutorial-keras-deep-learning-library/
for dna ony to int values
"""
def pad_n_repeat_sequences(sequences, maxlen=None, dtype='int32',
padding='post', truncating='post', value=0.):
"""extended version of pad_sequences()"""
if not hasattr(sequences, '__len__'):
raise ValueError('`sequences` must be iterable.')
lengths = []
for x in sequences:
if not hasattr(x, '__len__'):
raise ValueError('`sequences` must be a list of iterables. '
'Found non-iterable: ' + str(x))
lengths.append(len(x))
num_samples = len(sequences)
if maxlen is None:
maxlen = np.max(lengths) # sequences是基因序列,x是每一个序列的长度,求出最大值maxLen得到填充值
# take the sample shape from the first non empty sequence
# checking for consistency in the main loop below.
sample_shape = tuple()
for s in sequences:
if len(s) > 0:
sample_shape = np.asarray(s).shape[1:]
break
# make new array and fill with input seqs
x = (np.ones((num_samples, maxlen) + sample_shape) * value).astype(dtype) # np.ones((2, 1) + (1, 2)) 的 shape(2, 1, 1, 2)?? maybe 3维
for idx, s in enumerate(sequences):
if not len(s):
continue # empty list/array was found
if truncating == 'pre': # 加在序列的前面
trunc = s[-maxlen:]
elif truncating == 'post': # 加在序列的后面
trunc = s[:maxlen]
else:
raise ValueError('Truncating type "%s" not understood' % truncating)
# check `trunc` has expected shape
trunc = np.asarray(trunc, dtype=dtype)
if trunc.shape[1:] != sample_shape:
raise ValueError(
'Shape of sample %s of sequence at position %s is different from expected shape %s' %
(trunc.shape[1:], idx, sample_shape))
if repeat:
# repeat seq multiple times
repeat_seq = np.array([], dtype=dtype)
while len(repeat_seq) < maxLen:
if use_spacer:
spacer_length = random.randint(1, 50)
spacer = [value for i in range(spacer_length)]
repeat_seq = np.append(repeat_seq, spacer)
if randomrepeat:
random_start = random.randint(0, len(trunc))
repeat_seq = np.append(repeat_seq,
CircularList(trunc)[random_start:random_start + len(trunc)])
## 序列+间隔+序列
# 随机位置插入一段序列
else:
repeat_seq = np.append(repeat_seq, trunc)
else:
if randomrepeat:
random_start = random.randint(0, len(trunc))
repeat_seq = np.append(repeat_seq,
CircularList(trunc)[random_start:random_start + len(trunc)])
else:
repeat_seq = np.append(repeat_seq, trunc)
x[idx, :] = repeat_seq[-maxLen:]
else:
if padding == 'post':
x[idx, :len(trunc)] = trunc
elif padding == 'pre':
x[idx, -len(trunc):] = trunc
else:
raise ValueError('Padding type "%s" not understood' % padding)
return x
# ↑ 数据处理部分
encoder = LabelEncoder()
if len(x) > 0:
a = "ATGCN-"
encoder.fit(list(a)) # fit将a中的6个元素编码成0-5的数字
out = []
if type(x)==str:
dnaSeq = re.sub(r"[^ACGTUacgtu]", 'N', x)
encoded_X = encoder.transform(list(dnaSeq)) # transform将原始序列变成编码序列
out.append(encoded_X)
else:
for i in x:
dnaSeq = re.sub(r"[^ACGTUacgtu]", 'N', i)
# dnaSeq = i[0]
encoded_X = encoder.transform(list(dnaSeq))
out.append(encoded_X)
if online_Xtrain_set:
X_train_categorial = []
for seq in out:
X_train_categorial.append(np.array(to_categorical(seq, num_classes=len(a)), dtype=np.bool))
return X_train_categorial
else:
out = pad_n_repeat_sequences(out, maxlen=maxLen, dtype='int16', truncating='pre', value=0)
return np.array(to_categorical(out, num_classes=len(a)), dtype=np.bool)
else:
if y_encoder != None:
encoder.fit(y)
if np.array(encoder.classes_ != y_encoder.classes_).all():
warning(f"Warning not same classes in training and test set")
useable_classes = set(encoder.classes_).intersection(y_encoder.classes_) # 将X和Y放在一起
try:
assert np.array(encoder.classes_ == y_encoder.classes_).all()
except AssertionError:
warning(
f"not all test classes in training data, only {useable_classes} predictable "
f"from {len(encoder.classes_)} different classes\ntest set will be filtered so only predictable"
f" classes are included")
try:
assert len(useable_classes) == len(encoder.classes_) # 判断X和Y的类别长度是否相等
except AssertionError:
print(f"not all test classes in training data, only " \
f"{useable_classes} predictable from {len(encoder.classes_)} different classes" \
f"\ntest set will be filtered so only predictable classes are included")
if not len(useable_classes) == len(encoder.classes_):
global X_test, Y_test
arr = np.zeros(X_test.shape[0], dtype=int)
for i in useable_classes:
arr[y == i] = 1
X_test = X_test[arr == 1, :]
y = y[arr == 1]
encoded_Y = y_encoder.transform(y)
else:
encoded_Y = encoder.transform(y)
return to_categorical(encoded_Y, num_classes=len(y_encoder.classes_))
else:
encoder.fit(y)
# print(encoder.classes_)
# print(encoder.transform(encoder.classes_))
encoded_Y = encoder.transform(y)
return to_categorical(encoded_Y), encoder
先求出数据集中最长的基因序列的长度,构造一个新的array来承载新的处理过后的数据(用np.ones()初始化),在根据选项决定是否repeat、use spacer、random repeat以及决定padding的位置。在处理好基因序列填充之后,再将这些序列由字母转换成数字,再转换成one-hot编码。
如果采用online training的话,就直接输出原始数据进行微调的数据,否则输出原始数据经过基因填充处理过后的数据(107-115行)
最后对host文件的数据也进行了处理
def calc_shrink_size(seqlength):
subSeqlength = 100
for i in range(100, 400):
if (seqlength % i == 0):
subSeqlength = i
batch_size = int(seqlength / subSeqlength)
return subSeqlength, batch_size
def shrink_timesteps(X, Y, input_subSeqlength=0):
"""
needed for Truncated Backpropagation Through Time
If you have long input sequences, such as thousands of timesteps,
you may need to break the long input sequences into multiple contiguous subsequences.
e.g. 100 subseq.
Care would be needed to preserve state across each 100 subsequences and reset
the internal state after each 100 samples either explicitly or by using a batch size of 100.
:param input_subSeqlength: set for specific subsequence length
:return:
"""
# assert input_subSeqlength != 0, "must provide variable \"input_subSeqlength\" when using shrink_timesteps for specific subset"
if len(X.shape) == 3:
seqlength = X.shape[1]
features = X.shape[-1]
if input_subSeqlength == 0:
subSeqlength, batch_size = calc_shrink_size(seqlength)
else:
subSeqlength = input_subSeqlength
batch_size = int(seqlength / subSeqlength)
newSeqlength = int(seqlength / subSeqlength) * subSeqlength
bigarray = []
for sample in X:
sample = np.array(sample[0:newSeqlength], dtype=np.bool)
subarray = sample.reshape((int(seqlength / subSeqlength), subSeqlength, features))
bigarray.append(subarray)
bigarray = np.array(bigarray) # 把一个batch的数据拼接在一起
X = bigarray.reshape((bigarray.shape[0] * bigarray.shape[1], bigarray.shape[2], bigarray.shape[3]))
elif len(X.shape) == 2:
seqlength = X.shape[0]
features = X.shape[-1]
if input_subSeqlength == 0:
subSeqlength, batch_size = calc_shrink_size(seqlength)
else:
subSeqlength = input_subSeqlength
batch_size = int(seqlength / subSeqlength)
newSeqlength = int(seqlength / subSeqlength) * subSeqlength
sample = np.array(X[0:newSeqlength], dtype=np.bool)
subarray = sample.reshape((int(seqlength / subSeqlength), subSeqlength, features))
X = np.array(subarray)
else:
assert len(X.shape) == 2 or len(
X.shape) == 3, f"wrong shape of input X, expect len(shape) to be 2 or 3 but is instead {len(X.shape)}"
y = []
for i in Y:
y.append(int(seqlength / subSeqlength) * [i])
Y = np.array(y)
if len(Y.shape) == 2:
Y = np.array(y).flatten()
elif len(Y.shape) == 3:
Y = Y.reshape((Y.shape[0] * Y.shape[1], Y.shape[2]))
return X, Y, batch_size
如果基因序列过长(大于400),就要将过长的序列分割成为小段(100,400),在将一个batch的数据拼接在一起作为输入的数据。
数据生成器——DataGenerator.py
首先使用make_dataset.py来将数据按照比例划分成为X_train/val/test.csv以及Y_train/val/test.csv,make_dataset.py 比较简单故不再进行分析,直接使用即可。
def __data_generation(self, list_IDs_temp, indexes):
pool = multiprocessing.pool.ThreadPool()
'Generates data containing batch_size samples' # X : (n_samples, *dim, n_channels)
# Initialization
# X = np.empty((self.batch_size, self.dim, self.n_channels),dtype='str')
X = np.empty((self.number_samples_per_batch), dtype=object)
Y = np.empty((self.number_samples_per_batch), dtype=int)
sample_weight = np.array([])
def load_csv(sample):
X_i = pd.read_csv(os.path.join(self.directory, sample), delimiter='\t', dtype='str', header=None)[1].values[0]
return X_i
# Generate data
samples = pool.map(load_csv,list_IDs_temp)
X = np.array(samples)
for i, ID in enumerate(list_IDs_temp):
# Store sample
# load tsv, parse to numpy array, get str and set as value in X[i]
# X[i] = pd.read_csv(os.path.join(self.directory, ID), delimiter='\t', dtype='str', header=None)[1].values[0]
# sample_weight = np.append(sample_weight, 1)
# if len(X[i]) < self.dim:
# X[i] = "-" * self.dim
# sample_weight[i] = 0
# Store class
Y[i] = self.labels[indexes[i]]
sample_weight = np.array([[i] * self.number_subsequences for i in sample_weight]).flatten()
if self.maxLen == None:
maxLen = self.number_subsequences * self.dim
else:
maxLen = self.maxLen
# original_length = 50
# start_float = (original_length - self.sequence_length) / 2
# start = math.floor(start_float)
# stop = original_length - math.ceil(start_float)
# # amino = "GALMFWKQESPVICYHRNDTU"
# amino = "GALMFWKQESPVICYHRNDTUOBZX"
# encoder = LabelEncoder()
# encoder.fit(list(amino))
# X = parse_amino(x=[[i[start:stop]] for i in X], encoder=encoder)
# X = self.elmo_embedder.elmo_embedding(X, start, stop)
#
# X = seqvec.embed_sentence([i[start:stop] for i in X])
def encode_sample(sample):
X_i = DataParsing_main.encode_string(maxLen=maxLen, x=str(sample), repeat=self.repeat, use_spacer=self.use_spacer)
return X_i
X_wrong_shape = np.array(pool.map(encode_sample,X))
X = np.array(X_wrong_shape).reshape((X_wrong_shape.shape[0],-1,6))
# X = DataParsing.encode_string(maxLen=maxLen, x=X, repeat=self.repeat, use_spacer=self.use_spacer)
# assert self.shrink_timesteps != True or self.online_training != True, "online_training shrinks automatically " \
# "the files, please deactivate shrink_timesteps"
if self.online_training:
X, Y = DataParsing_main.manipulate_training_data(X=X, Y=Y, subSeqLength=self.dim,
number_subsequences=self.number_subsequences)
elif self.shrink_timesteps:
X, Y, batchsize = DataParsing_main.shrink_timesteps(input_subSeqlength=self.dim, X=X, Y=Y)
pool.close()
pool.join()
return X, tf.keras.utils.to_categorical(Y, num_classes=self.n_classes), sample_weight
加载CSV文件读取数据,以及其对应的类序号(不同的host是不同的类),得到最长基因序列长度,再用dataparsing的方法来处理数据
def _count_valid_files_in_directory(directory, white_list_formats, split,
follow_links):
"""
Copy from keras 2.1.5
Count files with extension in `white_list_formats` contained in directory.
Arguments:
directory: absolute path to the directory
containing files to be counted
white_list_formats: set of strings containing allowed extensions for
the files to be counted.
split: tuple of floats (e.g. `(0.2, 0.6)`) to only take into
account a certain fraction of files in each directory.
E.g.: `segment=(0.6, 1.0)` would only account for last 40 percent
of images in each directory.
follow_links: boolean.
Returns:
the count of files with extension in `white_list_formats` contained in
the directory.
"""
num_files = len(
list(_iter_valid_files(directory, white_list_formats, follow_links)))
if split:
start, stop = int(split[0] * num_files), int(split[1] * num_files)
else:
start, stop = 0, num_files
return stop - start
def parse_amino(x, encoder):
out = []
for i in x:
# dnaSeq = i[1].upper()
dnaSeq = i[0].upper()
encoded_X = encoder.transform(list(dnaSeq))
out.append(encoded_X)
return np.array(out)
def _list_valid_filenames_in_directory(directory, white_list_formats, split,
class_indices, follow_links):
"""Lists paths of files in `subdir` with extensions in `white_list_formats`.
Copy from keras-preprocessing 1.0.9
# Arguments
directory: absolute path to a directory containing the files to list.
The directory name is used as class label
and must be a key of `class_indices`.
white_list_formats: set of strings containing allowed extensions for
the files to be counted.
split: tuple of floats (e.g. `(0.2, 0.6)`) to only take into
account a certain fraction of files in each directory.
E.g.: `segment=(0.6, 1.0)` would only account for last 40 percent
of images in each directory.
class_indices: dictionary mapping a class name to its index.
follow_links: boolean.
# Returns
classes: a list of class indices
filenames: the path of valid files in `directory`, relative from
`directory`'s parent (e.g., if `directory` is "dataset/class1",
the filenames will be
`["class1/file1.jpg", "class1/file2.jpg", ...]`).
"""
dirname = os.path.basename(directory)
if split:
num_files = len(list(
_iter_valid_files(directory, white_list_formats, follow_links)))
start, stop = int(split[0] * num_files), int(split[1] * num_files)
valid_files = list(
_iter_valid_files(
directory, white_list_formats, follow_links))[start: stop]
else:
valid_files = _iter_valid_files(
directory, white_list_formats, follow_links)
classes = []
filenames = []
for root, fname in valid_files:
classes.append(class_indices[dirname])
absolute_path = os.path.join(root, fname)
relative_path = os.path.join(
dirname, os.path.relpath(absolute_path, directory))
filenames.append(relative_path)
return classes, filenames
def _iter_valid_files(directory, white_list_formats, follow_links):
"""Iterates on files with extension in `white_list_formats` contained in `directory`.
# Arguments
directory: Absolute path to the directory
containing files to be counted
white_list_formats: Set of strings containing allowed extensions for
the files to be counted.
follow_links: Boolean.
# Yields
Tuple of (root, filename) with extension in `white_list_formats`.
"""
def _recursive_list(subpath):
return sorted(os.walk(subpath, followlinks=follow_links),
key=lambda x: x[0]) # os.walk 列出一个地址的根目录 中间目录和文件名
for root, _, files in _recursive_list(directory):
for fname in sorted(files):
if fname.lower().endswith('.tiff'):
warnings.warn('Using ".tiff" files with multiple bands '
'will cause distortion. Please verify your output.')
if get_extension(fname) in white_list_formats:
yield root, fname
def get_extension(filename):
"""Get extension of the filename
There are newer methods to achieve this but this method is backwards compatible.
"""
return os.path.splitext(filename)[1].strip('.').lower()
读取文件目录中的文件,用生成器iterator输出,并且按照划分的比例来保留那些有效的文件
class DataGenerator(tf.keras.utils.Sequence):
'Generates data for Keras'
def __init__(self, directory, classes=None, number_subsequences=32, dim=(32, 32, 32), n_channels=6,
n_classes=10, shuffle=True, n_samples=None, seed=None, faster=True, online_training=False, repeat=True,
use_spacer=False, randomrepeat=False, sequence_length=50, number_samples_per_batch=32 , **kwargs):
'Initialization'
self.directory = directory
self.classes = classes
self.dim = dim
self.labels = None
self.list_IDs = None
self.n_channels = n_channels
self.shuffle = shuffle
self.seed = seed
self.online_training = online_training
self.repeat = repeat
self.use_spacer = use_spacer
self.randomrepeat = randomrepeat
self.maxLen = kwargs.get("maxLen", None)
self.sequence_length = sequence_length
if number_subsequences == 1:
self.shrink_timesteps = False ## 分割
else:
self.shrink_timesteps = True
self.number_subsequences = number_subsequences
if faster == True:
self.faster = 16
elif type(faster) == int and faster > 0:
self.faster = faster
else:
self.faster = 1
self.number_samples_per_batch = number_samples_per_batch * self.faster
self.number_samples_per_class_to_pick = n_samples
if not classes:
classes = []
for subdir in sorted(os.listdir(directory)):
if os.path.isdir(os.path.join(directory, subdir)):
classes.append(subdir)
self.classes = classes
self.n_classes = len(classes)
self.class_indices = dict(zip(classes, range(len(classes))))
# want a dict which contains dirs and number usable files
pool = multiprocessing.pool.ThreadPool()
function_partial = partial(_count_valid_files_in_directory,
white_list_formats={'csv'},
follow_links=None,
split=None) # partial 是用来冻结参数的,提供一个类似函数的方法
self.samples = pool.map(function_partial, (os.path.join(directory, subdir) for subdir in classes))
self.samples = dict(zip(classes, self.samples))
results = []
for dirpath in (os.path.join(directory, subdir) for subdir in classes):
results.append(pool.apply_async(_list_valid_filenames_in_directory,
(dirpath, {'csv'}, None, self.class_indices, None)))
# 使用apply_async即开始并行处理
self.filename_dict = {}
for res in results:
classes, filenames = res.get()
for index, class_i in enumerate(classes):
self.filename_dict.update({f"{class_i}_{index}": filenames[index]})
pool.close()
pool.join()
if not n_samples:
self.number_samples_per_class_to_pick = min(self.samples.values())
# self.elmo_embedder = Elmo_embedder()
self.elmo_embedder = None
self.on_epoch_end()
# in images wird ein groesses arr classes gemacht (fuer alle sampels) darin stehen OHE die Class
# erstelle filename liste in der die zugehoerige file adresse steht
# laesst sich mergen mit version die oben verlinked
def __len__(self):
'Denotes the number of batches per epoch'
return int(np.floor(len(self.list_IDs) / self.number_samples_per_batch))
def __getitem__(self, index):
'Generate one batch of data'
# Generate indexes of the batch
indexes = self.indexes[index * self.number_samples_per_batch:(index + 1) * self.number_samples_per_batch]
# Find list of IDs
list_IDs_temp = [self.list_IDs[k] for k in indexes]
# Generate data
X, y, sample_weight = self.__data_generation(list_IDs_temp, indexes)
return (X, y)
def on_epoch_end(self):
'make X-train sample list'
"""
1. go over each class
2. select randomly #n_sample samples of each class
3. add selection list to dict with class as key
"""
self.class_selection_path = np.array([])
self.labels = np.array([])
for class_i in self.classes:
samples_class_i = randsomsample(range(0, self.samples[class_i]), self.number_samples_per_class_to_pick)
self.class_selection_path = np.append(self.class_selection_path,
[self.filename_dict[f"{self.class_indices[class_i]}_{i}"] for i in
samples_class_i])
self.labels = np.append(self.labels, [self.class_indices[class_i] for i in samples_class_i])
self.list_IDs = self.class_selection_path
'Updates indexes after each epoch'
self.indexes = np.arange(len(self.list_IDs))
if self.shuffle == True:
if self.seed:
np.random.seed(self.seed)
np.random.shuffle(self.indexes)
本py文件主要的内容,用来在训练的每一个epoch生成一定class的一定的数据用做training,在每一个epoch end的时候,随机挑选下一个epoch所需要用到的sample,对应了文中的“随机重复欠采样”
training new model
def training(inpath, outpath, name, epochs, architecture, extention_variant, early_stopping, repeated_undersampling):
''' Train a model on your training files generated with make_dataset
\b
Example:
set input output and name of the model
$ vidhop train_new_model -i /home/user/trainingdata/ -o /home/user/model/ --name test
\b
use the LSTM archtecture and the extention variant random repeat
vidhop train_new_model -i /home/user/trainingdata/ --architecture 0 --extention_variant 2
\b
use repeated undersampling for training, note that for this the dataset must have been created with repeated undersampling enabled
vidhop train_new_model -i /home/user/trainingdata/ -r
\b
train the model for 40 epochs, stop training if for 2 epochs the accuracy did not increase
vidhop train_new_model -i /home/user/trainingdata/ --epochs 40 --early_stopping
'''
if extention_variant in (0, 1, 2, 3):
repeat = True
else:
repeat = False
if extention_variant in (2, 3):
randomrepeat = True
else:
randomrepeat = False
if extention_variant in (1, 3):
use_repeat_spacer = True
else:
use_repeat_spacer = False
if extention_variant == 5:
kwargs = dict({"maxLen": -1, "input_subSeqlength": 0})
else:
kwargs = dict()
if extention_variant == 6:
online_training = True
else:
online_training = False
if architecture == 0:
design = 4
else:
design = 7
files = os.listdir(inpath)
assert "Y_train.csv" in files, f"{inpath} must contain Y_train.csv file, but no such file in {files}"
test_and_plot(inpath=inpath, outpath=outpath, suffix=name, online_training=online_training, repeat=repeat,
randomrepeat=randomrepeat, early_stopping_bool=early_stopping, do_shrink_timesteps=True,
use_repeat_spacer=use_repeat_spacer, design=design, nodes=150, faster=True,
use_generator=repeated_undersampling, epochs=epochs, dropout=0.2, accuracy=True, **kwargs)
训练新的模型的主要方法,根据数据集调整的不同方法来输入相应的参数,从而可以修改模型输入数据集的构成。
class lrManipulator(tf.keras.callbacks.Callback):
"""
Manipulate the lr for Adam Optimizer
-> no big chances usefull
"""
def __init__(self, nb_epochs, nb_snapshots):
self.T = nb_epochs
self.M = nb_snapshots
def on_epoch_begin(self, epoch, logs={}):
tf.keras.backend.set_value(self.model.optimizer.lr, 0.001)
if ((epoch % (self.T // self.M)) == 0):
tf.keras.backend.set_value(self.model.optimizer.iterations, 0)
tf.keras.backend.set_value(self.model.optimizer.lr, 0.01)
class TimeHistory(tf.keras.callbacks.Callback): # 计时
"""https://stackoverflow.com/questions/43178668/record-the-computation-time-for-each-epoch-in-keras-during-model-fit"""
def on_train_begin(self, logs={}):
if not hasattr(self, 'times'):
self.times = []
self.time_train_start = time.time()
def on_epoch_end(self, batch, logs={}):
logs = logs or {}
self.times.append(int(time.time()) - int(self.time_train_start))
prediction_val = []
class accuracyHistory(tf.keras.callbacks.Callback):
"""to get the accuracy of my personal voting scores"""
def on_train_begin(self, logs={}):
if not hasattr(self, 'meanVote_val'):
self.meanVote_val = []
self.normalVote_val = []
def on_epoch_begin(self, epoch, logs=None):
global prediction_val
prediction_val = []
def on_epoch_end(self, batch, logs={}):
"""
1. make prediction of train
2. get the voting results
3. calc and save accuracy
4. do same for val set
"""
logs = logs or {}
global prediction_val
if (len(prediction_val) == 0):
prediction_val = (self.model.predict(X_val))
self.prediction_val = prediction_val
y_true_small, y_pred_mean_val, y_pred_voted_val, y_pred, y_pred_mean_exact = \
calc_predictions(X_val, Y_val, do_print=False, y_pred=self.prediction_val)
self.normalVote_val.append(metrics.accuracy_score(y_true_small, y_pred_voted_val))
self.meanVote_val.append(metrics.accuracy_score(y_true_small, y_pred_mean_val))
class roc_History(tf.keras.callbacks.Callback):
"""to get the AUC of my personal voting scores"""
# https://scikit-learn.org/stable/auto_examples/model_selection/plot_roc.html
def __init__(self, name, path):
self.name = name
self.path = path
def on_train_begin(self, logs={}):
if not hasattr(self, 'roc_val'):
# roc curve values for validation set
self.roc_macro = []
# roc curve values of the joined subsequences for the validation set
self.roc_mean_val = []
# roc curve values of the vote of the subsequences for the validation set
self.roc_meanVote_val = []
# thresholds per class
self.thresholds = []
# accuracy with general threshold tuning
self.acc_val_threshold_tuned = []
# accuracy with multi-threshold tuning
self.acc_val_multi_thresholds_tuned = []
def on_epoch_begin(self, epoch, logs=None):
global prediction_val
prediction_val = []
def on_epoch_end(self, batch, logs={}):
"""
1. make prediction of train
2. get the voting results
3. calc and save accuracy
4. do same for val set
"""
logs = logs or {}
# check if allready calculated validation results, if no calc new
global prediction_val
if (len(prediction_val) == 0):
prediction_val = (self.model.predict(X_val))
self.prediction_val = prediction_val
y_true_small, y_pred_mean_val, y_pred_voted_val, y_pred, y_pred_mean_val_exact = \
calc_predictions(X_val, Y_val, do_print=False, y_pred=self.prediction_val)
n_classes = Y_val.shape[-1]
y_true_small_bin = tf.keras.utils.to_categorical(y_true_small, n_classes)
y_pred_mean_val_bin = tf.keras.utils.to_categorical(y_pred_mean_val, n_classes)
调整学习率lr,记录、打印每个epoch所需要用到的时间、准确率以及AUC面积,AUC面积画图并输出(代码略)
class prediction_history(tf.keras.callbacks.Callback):
"""Callback subclass that prints each epoch prediction"""
def on_epoch_end(self, epoch, logs={}):
p = np.random.permutation(len(Y_val)) # 打乱顺序
shuffled_X = X_val[p]
shuffled_Y = Y_val[p]
self.predhis = (self.model.predict(shuffled_X[0:10]))
y_pred = np.argmax(self.predhis, axis=-1)
y_true = np.argmax(shuffled_Y, axis=-1)[0:10]
print(f"Predicted: {y_pred}")
print(f"True: {y_true}")
table = pd.crosstab(
pd.Series(y_true),
pd.Series(y_pred),
rownames=['True'],
colnames=['Predicted'],
margins=True)
print(table)
class History(tf.keras.callbacks.Callback):
"""
Callback that records events into a `History` object.
This callback is automatically applied to
every Keras model. The `History` object
gets returned by the `fit` method of models.
"""
def on_train_begin(self, logs=None):
if not hasattr(self, 'epoch'):
self.epoch = []
self.history = {}
def on_epoch_end(self, epoch, logs=None):
logs = logs or {}
self.epoch.append(epoch)
for k, v in logs.items():
self.history.setdefault(k, []).append(v)
class StopEarly(tf.keras.callbacks.Callback):
"""
Callback that stops training after an epoch
important for online training
"""
def on_epoch_end(self, epoch, logs=None):
self.model.stop_training = True
记录在val数据集上的predict准确率,记录历史数据,设置stopearly,即一定epoch内acc没有上升,则停止训练。
def model_for_plot(inpath, outpath, design=1, sampleSize=1, nodes=32, suffix="", epochs=100, dropout=0,
faster=False, voting=False, tensorboard=False, early_stopping_bool=True,
shuffleTraining=True, batch_norm=False, online_training=False,
number_subsequences=1, use_generator=True, repeat=True, use_spacer=False, randomrepeat=False,
**kwargs):
"""
method to train a model with specified properties, saves training behavior in /$path/"history"+suffix+".csv"
:param design: parameter for complexity of the NN, 0 == 2 layer GRU, 1 == 2 layer LSTM, 2 == 3 layer LSTM
:param sampleSize: fraction of samples that will be used for training (1/samplesize). 1 == all samples, 2 == half of the samples
:param nodes: number of nodes per layer
:param suffix: suffix for output files
:param epochs: number of epochs to train
:param dropout: rate of dropout to use, 0 == no Dropout, 0.2 = 20% Dropout
:param timesteps: size of "memory" of LSTM, don't change if not sure what you're doing
:param faster: speedup due higher batch size, can reduce accuracy
:param outpath: define the directory where the training history should be saved
:param voting: if true than saves the history of the voting / mean-predict subsequences, reduces training speed
:param tensorboard: for observing live changes to the network, more details see web
:param cuda: use GPU for calc, not tested jet, not working
:return: dict with loss and model
"""
model = tf.keras.models.Sequential()
global batch_size, X_train, X_test, Y_train
# Y_train_noOHE = np.argmax(Y_train, axis=1)
if use_generator:
class_weight = None
else:
Y_train_noOHE = [y.argmax() for y in Y_train]
class_weight = clw.compute_class_weight('balanced', np.unique(Y_train_noOHE), Y_train_noOHE)
class_weight_dict = {i: class_weight[i] for i in range(len(class_weight))}
class_weight = class_weight_dict
print(f"class_weights: {class_weight}")
timesteps = X_test.shape[1]
if faster:
batch = batch_size * 16
else:
batch = batch_size
if design == 0:
model.add(tf.keras.layers.GRU(nodes, input_shape=(timesteps, X_test.shape[-1]), return_sequences=True,
dropout=dropout))
model.add(tf.keras.layers.GRU(nodes, dropout=dropout))
if design == 1:
model.add(tf.keras.layers.LSTM(nodes, input_shape=(timesteps, X_test.shape[-1]), return_sequences=True,
dropout=dropout))
model.add(tf.keras.layers.LSTM(nodes, dropout=dropout))
if design == 2:
model.add(tf.keras.layers.LSTM(nodes, input_shape=(timesteps, X_test.shape[-1]), return_sequences=True,
dropout=dropout))
if batch_norm:
model.add(tf.keras.layers.BatchNormalization())
model.add(tf.keras.layers.LSTM(nodes, return_sequences=True, dropout=dropout))
if batch_norm:
model.add(tf.keras.layers.BatchNormalization())
model.add(tf.keras.layers.LSTM(nodes, dropout=dropout))
if batch_norm:
model.add(tf.keras.layers.BatchNormalization())
if design == 3:
model.add(tf.keras.layers.LSTM(nodes, input_shape=(timesteps, X_test.shape[-1]), return_sequences=True,
dropout=dropout))
model.add(tf.keras.layers.LSTM(nodes, return_sequences=True, dropout=dropout))
model.add(tf.keras.layers.LSTM(nodes, return_sequences=True, dropout=dropout))
model.add(tf.keras.layers.LSTM(nodes, dropout=dropout))
...... 省略部分代码
model.compile(optimizer='adam', loss='categorical_crossentropy', metrics=['acc'], sample_weight_mode=None)
# return model
filepath = outpath + "/model_best_acc_" + suffix + ".hdf5"
filepath2 = outpath + "/model_best_loss_" + suffix + ".hdf5"
checkpoint = tf.keras.callbacks.ModelCheckpoint(filepath, monitor='val_acc', verbose=1, save_best_only=True,
mode='max')
checkpoint2 = tf.keras.callbacks.ModelCheckpoint(filepath2, monitor='val_loss', verbose=1, save_best_only=True,
mode='min')
predictions = prediction_history()
time_callback = TimeHistory()
if early_stopping_bool:
early_stopping = tf.keras.callbacks.EarlyStopping('val_acc', min_delta=0, patience=epochs // 20,
restore_best_weights=True, verbose=2)
# early_stopping2 = EarlyStopping('val_loss', min_delta=0, patience=epochs//20,restore_best_weights=True)
callbacks_list = [checkpoint, checkpoint2, predictions, time_callback, early_stopping]
else:
callbacks_list = [checkpoint, checkpoint2, predictions, time_callback]
# callbacks_list = [early_stopping2, early_stopping, predictions, time_callback]
if voting:
myAccuracy = accuracyHistory()
myRoc = roc_History(name=suffix, path=outpath)
callbacks_list.append(myAccuracy)
callbacks_list.append(myRoc)
if tensorboard:
if not os.path.isdir(outpath + '/my_log_dir'):
os.makedirs(outpath + '/my_log_dir')
tensorboard = tf.keras.callbacks.TensorBoard(
# Log files will be written at this location
log_dir=outpath + '/my_log_dir',
# We will record activation histograms every 1 epoch
histogram_freq=1,
# We will record embedding data every 1 epoch
embeddings_freq=1,
)
tensorboard = tf.keras.callbacks.TensorBoard(log_dir=outpath + '/my_log_dir', histogram_freq=0, batch_size=32,
write_graph=True, write_grads=False, write_images=False,
embeddings_freq=0, embeddings_layer_names=None,
embeddings_metadata=None)
callbacks_list.append(tensorboard)
if use_generator:
from vidhop.training.DataGenerator import DataGenerator
params = {"number_subsequences": number_subsequences, "dim": timesteps, "n_channels": X_test.shape[-1],
"number_samples_per_batch": batch_size,
"n_classes": Y_test.shape[-1], "shuffle": shuffleTraining, "online_training": online_training,
"seed": 1, "repeat": repeat, "use_spacer": use_spacer, "randomrepeat": randomrepeat, "faster": faster}
# global directory
training_generator = DataGenerator(directory=inpath + "/train", **params, **kwargs)
hist = model.fit(training_generator, epochs=epochs, callbacks=callbacks_list, validation_data=(X_val, Y_val),
class_weight=class_weight, shuffle=shuffleTraining)
else:
if online_training == True:
print("use online training")
提供多种模型的组合来选择训练,从中可以对比出训练效果最好的模型,再对模型中的某些参数和选项进行调整,记录模型训练参数保存到文件里面去,
def calc_predictions(X, Y, y_pred, do_print=False):
"""
plot predictions
:param X: raw-data which should be predicted
:param Y: true labels for X
:param do_print: True == print the cross-tab of the prediction
:param y_pred: array with predicted labels for X
:return: y_true_small == True labels for complete sequences, yTrue == True labels for complete subsequences, y_pred_mean == with mean predicted labels for complete sequences, y_pred_voted == voted labels for complete sequences, y_pred == predicted labels for complete subsequences
"""
def print_predictions(y_true, y_pred, y_true_small, y_pred_voted, y_pred_sum, y_pred_mean_weight_std,
y_pred_mean_weight_ent):
table = pd.crosstab(
pd.Series(y_encoder.inverse_transform(y_true)),
pd.Series(y_encoder.inverse_transform(y_pred)),
rownames=['True'],
colnames=['Predicted'],
margins=True)
print("standard version")
print(table.to_string())
accuracy = metrics.accuracy_score(y_true, y_pred) * 100
print("standard version")
print("acc = " + str(accuracy))
table = pd.crosstab(
pd.Series(y_encoder.inverse_transform(y_true_small)),
pd.Series(y_encoder.inverse_transform(y_pred_voted)),
rownames=['True'],
colnames=['Predicted'],
margins=True)
print("vote version")
print(table.to_string())
accuracy = metrics.accuracy_score(y_true_small, y_pred_voted) * 100
print("vote version")
print("acc = " + str(accuracy))
table = pd.crosstab(
pd.Series(y_encoder.inverse_transform(y_true_small)),
pd.Series(y_encoder.inverse_transform(y_pred_sum)),
rownames=['True'],
colnames=['Predicted'],
margins=True)
······
根据文中列出的各种计算准确率的方法(vote, standard, std-div等)来计算准确率并输出对应的crosstable
model_path1 = f"{outpath}/model_best_loss_{suffix}.hdf5"
model_path2 = f"{outpath}/model_best_acc_{suffix}.hdf5"
for model_path in (model_path1, model_path2):
print("load model:")
print(model_path)
model = tf.keras.models.load_model(model_path)
pred = model.predict(X_test)
y_true_small, y_pred_mean, y_pred_voted, y_pred, y_pred_mean_exact = calc_predictions(X_test, Y_test,
y_pred=pred,
do_print=True)
print("make test")
myRoc = roc_History(name="_".join(model_path[len(outpath):].split("_")[1:3]) + "_" + suffix, path=outpath)
# myRoc = roc_History(name=suffix, path=outpath)
myRoc.on_train_begin()
global prediction_val
prediction_val = model.predict(X_test)
X_val = X_test
Y_val = Y_test
myRoc.on_epoch_end(0)
# create and export .model file
index_classes = dict()
for i in zip(y_encoder.transform(y_encoder.classes_), y_encoder.classes_):
index_classes.update({i[0]: i[1]})
repeat = True
use_spacer = False
online = False
random_repeat = True
design = design
multi_thresh = myRoc.thresholds[-1]
hosts = Y_test.shape[-1]
pickle.dump(
(model.to_json(), model.get_weights(), index_classes, multi_thresh, maxLen, repeat, use_repeat_spacer,
online_training, randomrepeat, design, hosts), open(f"{model_path.split('.hdf5')[0]}.model", "wb"))
从训练模型中获得的的最佳准确率对应的checkpoint还原模型,并且在text数据集上验证模型,得出 val acc 和 val loss。