OpenCV学习笔记(Python)———— 主动轮廓模型
本文包含主动轮廓模型代码以及实例分割代码
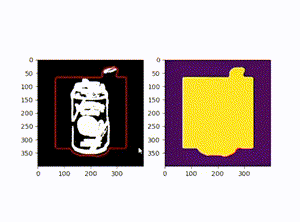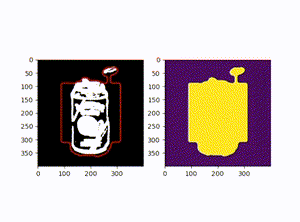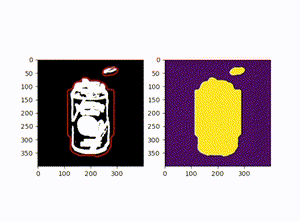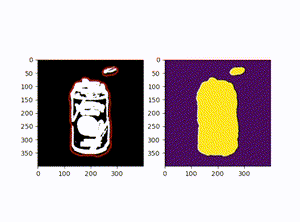
原图:
效果图:
主动轮廓模型: morphsnakes.py
# -*- coding: utf-8 -*-
"""
====================
Morphological Snakes
====================
"""
__author__ = "P. Márquez Neila "
from itertools import cycle
import numpy as np
from scipy import ndimage as ndi
__all__ = [
'morphological_chan_vese',
'morphological_geodesic_active_contour',
'inverse_gaussian_gradient',
'circle_level_set',
'checkerboard_level_set'
]
__version__ = (2, 1, 1)
__version_str__ = ".".join(map(str, __version__))
class _fcycle(object):
def __init__(self, iterable):
"""Call functions from the iterable each time it is called."""
self.funcs = cycle(iterable)
def __call__(self, *args, **kwargs):
f = next(self.funcs)
return f(*args, **kwargs)
# SI and IS operators for 2D and 3D.
_P2 = [np.eye(3),
np.array([[0, 1, 0]] * 3, dtype=object),
np.flipud(np.eye(3)),
np.rot90([[0, 1, 0]] * 3)]
_P3 = [np.zeros((3, 3, 3), dtype=object) for i in range(9)]
_P3[0][:, :, 1] = 1
_P3[1][:, 1, :] = 1
_P3[2][1, :, :] = 1
_P3[3][:, [0, 1, 2], [0, 1, 2]] = 1
_P3[4][:, [0, 1, 2], [2, 1, 0]] = 1
_P3[5][[0, 1, 2], :, [0, 1, 2]] = 1
_P3[6][[0, 1, 2], :, [2, 1, 0]] = 1
_P3[7][[0, 1, 2], [0, 1, 2], :] = 1
_P3[8][[0, 1, 2], [2, 1, 0], :] = 1
def sup_inf(u):
"""SI operator."""
if np.ndim(u) == 2:
P = _P2
elif np.ndim(u) == 3:
P = _P3
else:
raise ValueError("u has an invalid number of dimensions "
"(should be 2 or 3)")
erosions = []
for P_i in P:
erosions.append(ndi.binary_erosion(u, P_i))
return np.array(erosions, dtype=np.int8).max(0)
def inf_sup(u):
"""IS operator."""
if np.ndim(u) == 2:
P = _P2
elif np.ndim(u) == 3:
P = _P3
else:
raise ValueError("u has an invalid number of dimensions "
"(should be 2 or 3)")
dilations = []
for P_i in P:
dilations.append(ndi.binary_dilation(u, P_i))
return np.array(dilations, dtype=np.int8).min(0)
_curvop = _fcycle([lambda u: sup_inf(inf_sup(u)), # SIoIS
lambda u: inf_sup(sup_inf(u))]) # ISoSI
def _check_input(image, init_level_set):
"""Check that shapes of `image` and `init_level_set` match."""
if image.ndim not in [2, 3]:
raise ValueError("`image` must be a 2 or 3-dimensional array.")
if len(image.shape) != len(init_level_set.shape):
raise ValueError("The dimensions of the initial level set do not "
"match the dimensions of the image.")
def _init_level_set(init_level_set, image_shape):
if isinstance(init_level_set, str):
if init_level_set == 'checkerboard':
res = checkerboard_level_set(image_shape)
elif init_level_set == 'circle':
res = circle_level_set(image_shape)
elif init_level_set == 'ellipsoid':
res = ellipsoid_level_set(image_shape)
else:
raise ValueError("`init_level_set` not in "
"['checkerboard', 'circle', 'ellipsoid']")
else:
res = init_level_set
return res
def circle_level_set(image_shape, center=None, radius=None):
if center is None:
center = tuple(i // 2 for i in image_shape)
if radius is None:
radius = min(image_shape) * 3.0 / 8.0
grid = np.mgrid[[slice(i) for i in image_shape]]
grid = (grid.T - center).T
phi = radius - np.sqrt(np.sum((grid)**2, 0))
res = np.int8(phi > 0)
return res
def ellipsoid_level_set(image_shape, center=None, semi_axis=None):
if center is None:
center = tuple(i // 2 for i in image_shape)
if semi_axis is None:
semi_axis = tuple(i / 2 for i in image_shape)
if len(center) != len(image_shape):
raise ValueError("`center` and `image_shape` must have the same length.")
if len(semi_axis) != len(image_shape):
raise ValueError("`semi_axis` and `image_shape` must have the same length.")
if len(image_shape) == 2:
xc, yc = center
rx, ry = semi_axis
phi = 1 - np.fromfunction(
lambda x, y: ((x - xc) / rx) ** 2 +
((y - yc) / ry) ** 2,
image_shape, dtype=float)
elif len(image_shape) == 3:
xc, yc, zc = center
rx, ry, rz = semi_axis
phi = 1 - np.fromfunction(
lambda x, y, z: ((x - xc) / rx) ** 2 +
((y - yc) / ry) ** 2 +
((z - zc) / rz) ** 2,
image_shape, dtype=float)
else:
raise ValueError("`image_shape` must be a 2- or 3-tuple.")
res = np.int8(phi > 0)
return res
def checkerboard_level_set(image_shape, square_size=5):
grid = np.ogrid[[slice(i) for i in image_shape]]
grid = [(grid_i // square_size) & 1 for grid_i in grid]
checkerboard = np.bitwise_xor.reduce(grid, axis=0, )
res = np.int8(checkerboard)
return res
def inverse_gaussian_gradient(image, alpha=100.0, sigma=5.0):
gradnorm = ndi.gaussian_gradient_magnitude(image, sigma, mode='nearest')
return 1.0 / np.sqrt(1.0 + alpha * gradnorm)
def morphological_chan_vese(image, iterations, init_level_set='checkerboard',
smoothing=1, lambda1=1, lambda2=1,
iter_callback=lambda x: None):
init_level_set = _init_level_set(init_level_set, image.shape)
_check_input(image, init_level_set)
u = np.int8(init_level_set > 0)
iter_callback(u)
for _ in range(iterations):
# inside = u > 0
# outside = u <= 0
c0 = (image * (1 - u)).sum() / float((1 - u).sum() + 1e-8)
c1 = (image * u).sum() / float(u.sum() + 1e-8)
# Image attachment
du = np.gradient(u)
abs_du = np.abs(du).sum(0)
aux = abs_du * (lambda1 * (image - c1)**2 - lambda2 * (image - c0)**2)
u[aux < 0] = 1
u[aux > 0] = 0
# Smoothing
for _ in range(smoothing):
u = _curvop(u)
iter_callback(u)
return u
def morphological_geodesic_active_contour(gimage , iterations,
init_level_set='circle', smoothing=1,
threshold='auto', balloon=0,
iter_callback=lambda x: None):
image = gimage
init_level_set = _init_level_set(init_level_set, image.shape)
_check_input(image, init_level_set)
if threshold == 'auto':
threshold = np.percentile(image, 40)
structure = np.ones((3,) * len(image.shape), dtype=np.int8)
dimage = np.gradient(image)
# threshold_mask = image > threshold
if balloon != 0:
threshold_mask_balloon = image > threshold / np.abs(balloon)
u = np.int8(init_level_set > 0)
iter_callback(u)
for _ in range(iterations):
# Balloon
if balloon > 0:
aux = ndi.binary_dilation(u, structure)
elif balloon < 0:
aux = ndi.binary_erosion(u, structure)
if balloon != 0:
u[threshold_mask_balloon] = aux[threshold_mask_balloon]
# Image attachment
aux = np.zeros_like(image)
du = np.gradient(u)
for el1, el2 in zip(dimage, du):
aux += el1 * el2
u[aux > 0] = 1
u[aux < 0] = 0
# Smoothing
for _ in range(smoothing):
u = _curvop(u)
iter_callback(u)
return u
例子代码:
import logging
import cv2
import numpy as np
from imageio import imread
import matplotlib
from matplotlib import pyplot as plt
import morphsnakes as ms # 调用morphsnakes.py
matplotlib.use('TKAgg') # 在TKAgg上运行
def visual_callback_2d(background, fig=None):
# Prepare the visual environment.
if fig is None:
fig = plt.figure()
fig.clf()
ax1 = fig.add_subplot(1, 2, 1)
ax1.imshow(background, cmap=plt.cm.gray)
ax2 = fig.add_subplot(1, 2, 2)
ax_u = ax2.imshow(np.zeros_like(background), vmin=0, vmax=1)
plt.pause(0.001)
def callback(levelset):
if ax1.collections:
del ax1.collections[0]
ax1.contour(levelset, [0.5], colors='r')
ax_u.set_data(levelset)
fig.canvas.draw()
plt.pause(0.001)
return callback
def example(PATH):
logging.info('Running: example_coins (MorphGAC)...')
# Load the image.
imgcolor = imread(PATH)
# 梯度 = 膨胀 - 腐蚀
kernel = np.ones((8, 8), np.uint8)
cv2.dilate(imgcolor, kernel, iterations=5) # 膨胀
cv2.erode(imgcolor, kernel, iterations=5) # 腐蚀
gradient = cv2.morphologyEx(imgcolor, cv2.MORPH_GRADIENT, kernel)
gray = cv2.cvtColor(gradient, cv2.COLOR_RGB2GRAY) # 转化为灰度图像
ret, img = cv2.threshold(gray, 127, 255, cv2.THRESH_BINARY) # 二化值 根据明暗调节第二个值
# g(I)
gimg = ms.inverse_gaussian_gradient(img, alpha=500, sigma=4.0) # 反高斯梯度
# 初始轮廓设置
init_ls = np.zeros(img.shape, dtype=np.int8)
init_ls[10:-10, 10:-10] = 1
# Callback for visual plotting
callback = visual_callback_2d(img)
# MorphGAC.
ms.morphological_geodesic_active_contour(gimg, 100, init_ls,
smoothing=5, threshold=0.1,
balloon=-1, iter_callback=callback)
if __name__ == '__main__':
Path = r"E:\Water_bottle_small.jpg" # 将这里换成你自己的图片路径
example(Path)
plt.show()