机器学习之泰坦尼克号实战
记录一下利用Python线性回归、逻辑回归、随机森林算法处理泰坦尼克号数据的过程,对比准确率。
Data Preprocessing
先打出来5行看看数据
import pandas
titanic = pandas.read_csv("titanic_train.csv")
titanic.head(5)
titanic["Age"] = titanic["Age"].fillna(titanic["Age"].median())
titanic.head()

对于不是纯数字的列,将字符或字符串换成数字,如将Sex列的性别划分0和1
print (titanic["Sex"].unique()) #看有几个不一样
#换0 1
# Replace all the occurences of male with the number 0.
titanic.loc[titanic["Sex"] == "male", "Sex"] = 0
titanic.loc[titanic["Sex"] == "female", "Sex"] = 1
同时,对于字符列,谁出现的多,就把NaN值换成谁
#字符型的有NaN值谁多用谁
#换0 1 2
print (titanic["Embarked"].unique())
titanic["Embarked"] = titanic["Embarked"].fillna('S')
titanic.loc[titanic["Embarked"] == "S", "Embarked"] = 0
titanic.loc[titanic["Embarked"] == "C", "Embarked"] = 1
titanic.loc[titanic["Embarked"] == "Q", "Embarked"] = 2
利用Linear Regression
# Import the linear regression class
from sklearn.linear_model import LinearRegression
# Sklearn also has a helper that makes it easy to do cross validation
from sklearn.model_selection import KFold
# The columns we'll use to predict the target
predictors = ["Pclass", "Sex", "Age", "SibSp", "Parch", "Fare", "Embarked"]
# Initialize our algorithm class
alg = LinearRegression()
# Generate cross validation folds for the titanic dataset. It return the row indices corresponding to train and test.
# We set random_state to ensure we get the same splits every time we run this.
#n_folds变为n_splits
kf = KFold(titanic.shape[0], n_splits=3, random_state=1, shuffle=False)
predictions = []
for train, test in kf.split(titanic):
# The predictors we're using the train the algorithm. Note how we only take the rows in the train folds.
train_predictors = (titanic[predictors].iloc[train,:])
# The target we're using to train the algorithm.
train_target = titanic["Survived"].iloc[train]
# Training the algorithm using the predictors and target.
alg.fit(train_predictors, train_target)
# We can now make predictions on the test fold
test_predictions = alg.predict(titanic[predictors].iloc[test,:])
predictions.append(test_predictions)
已经建立了模型,得到获救的值,现在要把这个值换成是否获救(0/1),再看下线性回归的精度
import numpy as np
# The predictions are in three separate numpy arrays. Concatenate them into one.
# We concatenate them on axis 0, as they only have one axis.
predictions = np.concatenate(predictions, axis=0)
# Map predictions to outcomes (only possible outcomes are 1 and 0)
predictions[predictions > .5] = 1
predictions[predictions <=.5] = 0
######################
#这里不太确定,是用sum还是len
#accuracy = sum(predictions[predictions == titanic["Survived"]]) / len(predictions)
#accuracy = predictions[predictions == titanic["Survived"]].size / len(predictions)
accuracy = sum(predictions == titanic["Survived"]) / len(predictions)
print (accuracy)
0.7968574635241302
利用Logistics Regression
#from sklearn import cross_validation 旧库已经不行了,这里要注意
from sklearn import model_selection
from sklearn.linear_model import LogisticRegression
# Initialize our algorithm
alg = LogisticRegression(random_state=1)
# Compute the accuracy score for all the cross validation folds. (much simpler than what we did before!)
scores = model_selection.cross_val_score(alg, titanic[predictors], titanic["Survived"], cv=3)
# Take the mean of the scores (because we have one for each fold)
print(scores.mean())
0.7957351290684623
在测试集试试
同样如训练集中的操作
titanic_test = pandas.read_csv("test.csv")
titanic_test["Age"] = titanic_test["Age"].fillna(titanic["Age"].median())
titanic_test["Fare"] = titanic_test["Fare"].fillna(titanic_test["Fare"].median())
titanic_test.loc[titanic_test["Sex"] == "male", "Sex"] = 0
titanic_test.loc[titanic_test["Sex"] == "female", "Sex"] = 1
titanic_test["Embarked"] = titanic_test["Embarked"].fillna("S")
titanic_test.loc[titanic_test["Embarked"] == "S", "Embarked"] = 0
titanic_test.loc[titanic_test["Embarked"] == "C", "Embarked"] = 1
titanic_test.loc[titanic_test["Embarked"] == "Q", "Embarked"] = 2
利用模型计算部分都一样,先挖个坑。
利用Random Forest
一般树的高度不要太高,否则会过拟合
from sklearn import model_selection
from sklearn.ensemble import RandomForestClassifier
predictors = ["Pclass", "Sex", "Age", "SibSp", "Parch", "Fare", "Embarked"]
# Initialize our algorithm with the default paramters
# n_estimators is the number of trees we want to make
# min_samples_split is the minimum number of rows we need to make a split
# min_samples_leaf is the minimum number of samples we can have at the place where a tree branch ends (the bottom points of the tree)
#10:树的个数 2:当样本数是2的时候不切 1:叶子节点个数最少1个
alg = RandomForestClassifier(random_state=1, n_estimators=10, min_samples_split=2, min_samples_leaf=1)
# Compute the accuracy score for all the cross validation folds. (much simpler than what we did before!)
kf = model_selection.KFold(titanic.shape[0], n_splits=3, random_state=1)
scores = model_selection.cross_val_score(alg, titanic[predictors], titanic["Survived"], cv=kf)
# Take the mean of the scores (because we have one for each fold)
print(scores.mean())
0.8069584736251403
然后是最重要的参数调节,可以调n_estimators, min_samples_split, min_samples_leaf这三个参数,调节后两个参数可以调整树的高度,第一个参数可以调树的个数,10个树不够,可以换50个、100个。
alg = RandomForestClassifier(random_state=1, n_estimators=50, min_samples_split=4, min_samples_leaf=2)
# Compute the accuracy score for all the cross validation folds. (much simpler than what we did before!)
kf = model_selection.KFold(titanic.shape[0], n_splits=3, random_state=1)
scores = model_selection.cross_val_score(alg, titanic[predictors], titanic["Survived"], cv=kf)
# Take the mean of the scores (because we have one for each fold)
print(scores.mean())
0.8260381593714927
最重要的就是参数调优!此时可以看到准确率有提升。还想提准确率,此时回到数据本身,再加几个特征:SibSp、Parch。再考虑一个玄学的东西:Name,名字长短可能跟活下来有关…如果无关可以去掉,先加入考虑范围。
#提取特征
# Generating a familysize column
titanic["FamilySize"] = titanic["SibSp"] + titanic["Parch"]
# The .apply method generates a new series
titanic["NameLength"] = titanic["Name"].apply(lambda x: len(x))
把称谓里面的Dr、Mr、Mrs提取出来
import re
# A function to get the title from a name.
def get_title(name):
# Use a regular expression to search for a title. Titles always consist of capital and lowercase letters, and end with a period.
title_search = re.search(' ([A-Za-z]+)\.', name)
# If the title exists, extract and return it.
if title_search:
return title_search.group(1)
return ""
# Get all the titles and print how often each one occurs.
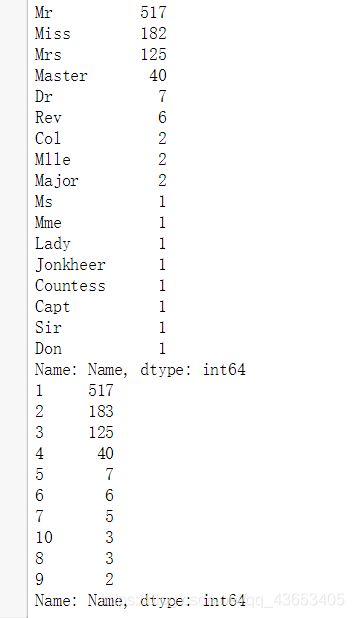
titles = titanic["Name"].apply(get_title)
print(pandas.value_counts(titles))
# Map each title to an integer. Some titles are very rare, and are compressed into the same codes as other titles.
title_mapping = {"Mr": 1, "Miss": 2, "Mrs": 3, "Master": 4, "Dr": 5, "Rev": 6, "Major": 7, "Col": 7, "Mlle": 8, "Mme": 8, "Don": 9, "Lady": 10, "Countess": 10, "Jonkheer": 10, "Sir": 9, "Capt": 7, "Ms": 2}
for k,v in title_mapping.items():
titles[titles == k] = v
# Verify that we converted everything.
print(pandas.value_counts(titles))
# Add in the title column.
titanic["Title"] = titles
那么多特征,看一下哪个特征比较重要,用到python自带的包:feature_selection。
import numpy as np
from sklearn.feature_selection import SelectKBest, f_classif
import matplotlib.pyplot as plt
predictors = ["Pclass", "Sex", "Age", "SibSp", "Parch", "Fare", "Embarked", "FamilySize", "Title", "NameLength"]
# Perform feature selection
selector = SelectKBest(f_classif, k=5)
selector.fit(titanic[predictors], titanic["Survived"])
# Get the raw p-values for each feature, and transform from p-values into scores
scores = -np.log10(selector.pvalues_)
# Plot the scores. See how "Pclass", "Sex", "Title", and "Fare" are the best?
plt.bar(range(len(predictors)), scores)
plt.xticks(range(len(predictors)), predictors, rotation='vertical')
plt.show()
# Pick only the four best features.
predictors = ["Pclass", "Sex", "Fare", "Title"]
alg = RandomForestClassifier(random_state=1, n_estimators=50, min_samples_split=8, min_samples_leaf=4)
谁高谁的谁就更重要。此时可以选出特征:Pclass、Sex、Fare、Title、NameLength。
多个分类器一起上,最后求均值
竞赛和论文的神器。。。
from sklearn.ensemble import GradientBoostingClassifier
import numpy as np
# The algorithms we want to ensemble.
# We're using the more linear predictors for the logistic regression, and everything with the gradient boosting classifier.
algorithms = [
[GradientBoostingClassifier(random_state=1, n_estimators=25, max_depth=3), ["Pclass", "Sex", "Age", "Fare", "Embarked", "FamilySize", "Title",]],
[LogisticRegression(random_state=1), ["Pclass", "Sex", "Fare", "FamilySize", "Title", "Age", "Embarked"]]
]
# Initialize the cross validation folds
kf = KFold(titanic.shape[0], n_splits=3, random_state=1)
predictions = []
for train, test in kf.split(titanic):
train_target = titanic["Survived"].iloc[train]
full_test_predictions = []
# Make predictions for each algorithm on each fold
for alg, predictors in algorithms:
# Fit the algorithm on the training data.
alg.fit(titanic[predictors].iloc[train,:], train_target)
# Select and predict on the test fold.
# The .astype(float) is necessary to convert the dataframe to all floats and avoid an sklearn error.
test_predictions = alg.predict_proba(titanic[predictors].iloc[test,:].astype(float))[:,1]
full_test_predictions.append(test_predictions)
# Use a simple ensembling scheme -- just average the predictions to get the final classification.
test_predictions = (full_test_predictions[0] + full_test_predictions[1]) / 2
# Any value over .5 is assumed to be a 1 prediction, and below .5 is a 0 prediction.
test_predictions[test_predictions <= .5] = 0
test_predictions[test_predictions > .5] = 1
predictions.append(test_predictions)
# Put all the predictions together into one array.
predictions = np.concatenate(predictions, axis=0)
# Compute accuracy by comparing to the training data.
accuracy = sum(predictions[predictions == titanic["Survived"]]) / len(predictions)
print(accuracy)