脑电数据预处理和后续处理(EEGLAB)
目录
-
- EEGLAB
-
- 下载与安装
- 载入数据
- 绘制脑地形图
- 脑电数据预处理
-
- 参考转换(Re-reference)
- 滤波
- 改变采样率
- 移除无用电极
- 分段
- 基线校正
- 伪迹去除
- 剔除坏段
- 叠加平均
- 保存数据
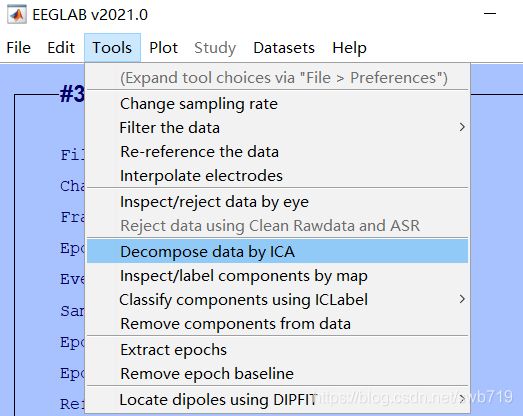
- 脑电分类
-
- 特征提取
- 分类
由于我的毕设涉及到脑电信号的处理与分类,所以在这记录一下我的学习过程。不得不说我留给毕设的时间好像不足以支撑到我完全理解eeglab的使用…
EEGLAB
下载与安装
我的matlab版本是R2018a,在eeglab官网下载了之后放到了matlab文件夹下toolbox里边,然后再添加路径就可以使用了。
出现的问题
当打开eeglab时,界面提醒 R2018a需要升级,同时eeglab运行到关掉某个进行过程时,matlab会崩溃
然后我下载了R2018a升级的压缩包,解决了这个问题。
载入数据
- 采集之后我得到的数据是cnt的文件,在界面的file–>import data下可以选择导入cnt格式的数据,如下图:
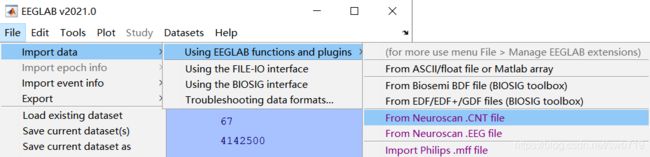
载入数据之后,可以看到界面显示为:

这里我出现了个问题,我采集脑电设备的电极是64极的,但是eeglab显示是67个通道,其中多了三个通道,观察数据发现有三行值是100到500多的,搞不懂为什么,如果有知道的朋友希望评论一下,谢谢 我感觉这个问题是个雷,可能之后的处理操作就因为这个都错了…
更新:
参考:link
这个网页中描述的问题和我的相似,他采集设备的电极是128,但是导入后eeglab显示有132个通道。回答说FidNz = the fiducial marker near the nose (nasion),eeglab会自动检测出三个隐藏通道,然后建议是选择比采集电极多的电极位置图,不过也要保证电极位置的正确。 - plot–>channel data(scroll) 可以观察对应电极通道的序列图。
绘制脑地形图
edit --> channel location --> read locations --> 选择位置文件
这里需要注意的是如果要选择位置文件的话(也可以不选择,eeglab有自动匹配),在eeglab的文件夹中的sample_locs下可以选择对应的文件。被试佩戴脑电帽的通道数为 64 导,电极位置遵照国际 10-20 标准分布。
因为我没有找到对应的电极位置文件,所以我并没有选择 Read Location,直接用的eeglab自动对准的。
脑电数据预处理
参考转换(Re-reference)
滤波
这部分具体应该选择的频率范围我不是很确定,我选择了0.25~30Hz,依次在第一框中填入0.25和第二个框中填入30进行计算,两个一起填代表进行带通滤波,建议分开进行,先高通滤波再低通滤波。

改变采样率
原数据的采样率是1000Hz,很占内存,所以我将采样率改为120Hz。
降低采样率需要注意:
- 一是降低采样率要在滤波之后。因为降低采样率会使我们丢失高频信息,使高频信息变得扭曲,所以最好在保留了我们感兴趣波段之后再去降低采样率,这样可以保证信号最大程度不会失真。
- 二是根据负荷采样定理:选择是我们想要分析的波段频率的3,4倍以上,理论上来说,我们的采样率必须是我们想要分析的波段的两倍,比如我们想要分析60Hz的波,那数据采样率为120Hz就足够了。实际上,建议采样率最好在分析波段的三到四倍。
移除无用电极
分段

一般的ERP分段在事件前200毫秒到事件后1000毫秒这个时间段就足够了,因为几乎所有的ERP成分,都是在1秒以内产生的。但是如果后期还要做时频分析的话,则需要将分段时间拉长,扩大到事件前1秒到事件后2秒这个时间段内,因为时频分析的算法,要求数据要有一定的长度。
基线校正
分段之后就会跳出基线校正的操作框。
伪迹去除
绘制2-D Component Scalp Maps,这一步需要对每一个成分进行查看和判断,然后将想要剔除的成分先标记起来。

剔除ICA成分

剔除哪些成分,我是借助了Adjust插件(Adjust 需要自己安装,不是 EEGLAB 默认携带的。)
绘制 component spectra and maps,了解哪些独立成分对特定的频段贡献最大,这里我设置需要随机选择的数据的百分比为100%。
command line上显示的结果如下:
Component 1 percent relative variance: 83.77
Component 2 percent relative variance: 12.14
Component 3 percent relative variance: 6.57
Component 4 percent relative variance: 7.76
Component 5 percent relative variance: 8.08
Component 6 percent relative variance: 14.21
Component 7 percent relative variance: 2.81
Component 8 percent relative variance: 6.20
Component 9 percent relative variance: 4.55
Component 10 percent relative variance: 1.20
Component 11 percent relative variance: 3.25
Component 12 percent relative variance: 2.63
Component 13 percent relative variance: 1.39
Component 14 percent relative variance: 2.48
Component 15 percent relative variance: 2.11
Component 16 percent relative variance: 1.41
Component 17 percent relative variance: 1.35
Component 18 percent relative variance: 1.64
Component 19 percent relative variance: 0.24
Component 20 percent relative variance: 0.82
Component 21 percent relative variance: 0.38
Component 22 percent relative variance: 0.60
Component 23 percent relative variance: 0.89
Component 24 percent relative variance: 1.27
Component 25 percent relative variance: 0.44
Component 26 percent relative variance: 0.54
Component 27 percent relative variance: 0.61
Component 28 percent relative variance: 0.22
Component 29 percent relative variance: 0.45
Component 30 percent relative variance: 0.32
Component 31 percent relative variance: 1.13
Component 32 percent relative variance: 0.09
Component 33 percent relative variance: 0.67
Component 34 percent relative variance: 0.22
Component 35 percent relative variance: 0.21
Component 36 percent relative variance: 0.28
Component 37 percent relative variance: 0.50
Component 38 percent relative variance: 0.16
Component 39 percent relative variance: 0.26
Component 40 percent relative variance: 0.14
Component 41 percent relative variance: 0.18
Component 42 percent relative variance: 0.18
Component 43 percent relative variance: 0.07
Component 44 percent relative variance: 0.02
Component 45 percent relative variance: 0.17
Component 46 percent relative variance: 0.05
Component 47 percent relative variance: 0.02
Component 48 percent relative variance: 0.10
Component 49 percent relative variance: 0.04
Component 50 percent relative variance: 0.02
Component 51 percent relative variance: 0.01
Component 52 percent relative variance: 0.00
Component 53 percent relative variance: 0.01
Component 54 percent relative variance: 0.01
Component 55 percent relative variance: 0.00
Component 56 percent relative variance: 0.00
Component 57 percent relative variance: 0.00
为了得到独立成分的 ERP 对数据的 ERP 的贡献,我们需要选择 Plot > Component ERPs> with component maps。
command line上显示:
Comparing maximum projections for components:
IC1 maximum mean power of back-projection: 72.2738
IC2 maximum mean power of back-projection: 40.3074
IC3 maximum mean power of back-projection: 118.629
IC4 maximum mean power of back-projection: 20.5741
IC5 maximum mean power of back-projection: 16.7698
IC6 maximum mean power of back-projection: 4.04828
IC7 maximum mean power of back-projection: 5.03374
IC8 maximum mean power of back-projection: 27.3447
IC9 maximum mean power of back-projection: 12.9047
IC10 maximum mean power of back-projection: 2.4831
IC11 maximum mean power of back-projection: 5.01077
IC12 maximum mean power of back-projection: 3.94776
IC13 maximum mean power of back-projection: 5.80443
IC14 maximum mean power of back-projection: 10.3948
IC15 maximum mean power of back-projection: 11.8145
IC16 maximum mean power of back-projection: 1.14692
IC17 maximum mean power of back-projection: 2.63034
IC18 maximum mean power of back-projection: 4.09341
IC19 maximum mean power of back-projection: 2.35853
IC20 maximum mean power of back-projection: 1.54849
IC21 maximum mean power of back-projection: 0.761995
IC22 maximum mean power of back-projection: 1.22335
IC23 maximum mean power of back-projection: 1.50031
IC24 maximum mean power of back-projection: 6.12206
IC25 maximum mean power of back-projection: 0.449215
IC26 maximum mean power of back-projection: 1.05872
IC27 maximum mean power of back-projection: 0.574225
IC28 maximum mean power of back-projection: 0.44772
IC29 maximum mean power of back-projection: 0.545843
IC30 maximum mean power of back-projection: 0.624281
IC31 maximum mean power of back-projection: 1.91398
IC32 maximum mean power of back-projection: 0.356062
IC33 maximum mean power of back-projection: 0.483526
IC34 maximum mean power of back-projection: 0.403002
IC35 maximum mean power of back-projection: 0.166475
IC36 maximum mean power of back-projection: 0.393186
IC37 maximum mean power of back-projection: 0.468765
IC38 maximum mean power of back-projection: 0.336584
IC39 maximum mean power of back-projection: 0.495862
IC40 maximum mean power of back-projection: 0.29598
IC41 maximum mean power of back-projection: 0.115158
IC42 maximum mean power of back-projection: 0.191568
IC43 maximum mean power of back-projection: 0.0438948
IC44 maximum mean power of back-projection: 0.073342
IC45 maximum mean power of back-projection: 0.0760424
IC46 maximum mean power of back-projection: 0.238884
IC47 maximum mean power of back-projection: 0.0109103
IC48 maximum mean power of back-projection: 0.0265037
IC49 maximum mean power of back-projection: 0.016949
IC50 maximum mean power of back-projection: 0.00671989
IC51 maximum mean power of back-projection: 0.00366287
IC52 maximum mean power of back-projection: 22.7095
IC53 maximum mean power of back-projection: 0.00337244
IC54 maximum mean power of back-projection: 0.00276348
IC55 maximum mean power of back-projection: 0.00112066
IC56 maximum mean power of back-projection: 0.000727603
IC57 maximum mean power of back-projection: 0.000860832
in the interval -1000 ms to 1992 ms.
Plotting envelopes of 7 component projections.
Topo maps will show components: 52 5 2 8 4 1 3
with max var at times (ms): 8 242 675 692 1617 1750 1783
epoch frames: 122 150 202 204 315 331 335
Component sortvar in interval: 72.27 40.31 118.63 20.57 16.77 4.05 5.03
Summed component 'ppaf' in interval [-1000 1991.67] ms: 53.83%
Plot limits (sec, sec, uV, uV) [-1,1.99167,-20.1883,13.5947]
绘制Component ERP-image
剔除坏段
最后一个步骤就是要剔除掉波幅超过100微幅的trial(要求严格一点也可以定位80),因为波幅起伏这么大的活动,已经不是由认知活动引起的了,肯定是由于其他的动作引起的,所以可以直接剔除掉。
这一步操作一定要在插值坏导和ICA去除眼电成分之后再做,因为有可能被试眨眼比较频繁,那这么操作下来,有可能就需要删掉接近一半的trials数了,删掉的trials数太多,这个数据也就废了。所以要将它留到最后一步才做,同时,这个操作也能用来检查我们前两步操作是否足够有效。
我采用的是绝对阈值法,选择剔除掉超出±100μV的trial。
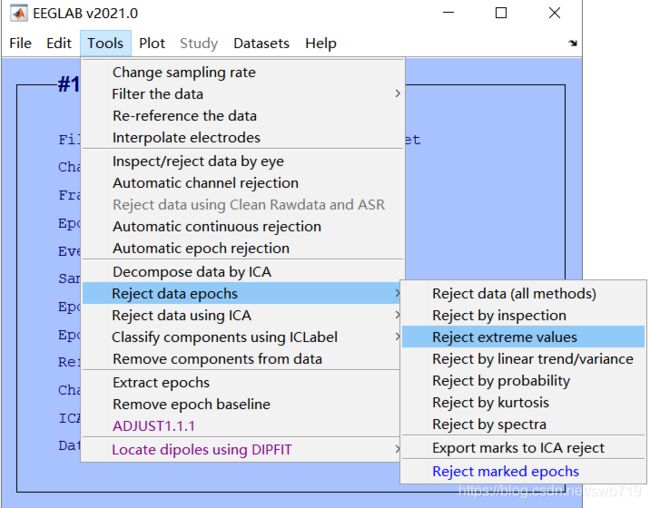
2020版本的eeglab中默认没有显示reject data,file>preference中可以设置用以前版本的GUI界面,如下图:

然后选择reject data epochs>reject extreme values
command line显示:
250/540 trials marked for rejection
也太多了吧…这就要删掉近一半的trial了,那我还是观察时域波形,手动删除不良片段吧。如何确定坏段呢?
叠加平均
保存数据
脑电分类
起初我一直在纠结的是我脑电数据对应的标签要怎么弄,后来我明白了分段之后EEG数据下有个epoch的字段就是事件类型。
eeglab中可以看到数据的event type,截图如下:第一位代表的是变量一的标签,第二位表示变量二的标签,这个取决于脑电采集过程中的打标设置(255好像是个意外)。其余部分后续补上。

特征提取
分类
- 划分训练集和测试集
- 调库,选择多种分类器训练以及测试
参考:
https://blog.csdn.net/zyb228/article/details/106216744
https://www.jianshu.com/p/0df08244614a
https://roses.blog.csdn.net/article/details/106216744