(单细胞-SingleCell)Scanpy流程——python 实现单细胞 Seurat 流程——单细胞文献实战
导入模块
import scanpy as sc
import os
import math
import itertools
import warnings
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
%config InlineBackend.figure_format = 'svg'
warnings.filterwarnings("ignore")
plt.rc('font',family='Times New Roman')
my_colors = ["#1EB2A6","#ffc4a3","#e2979c","#F67575"]
sc.settings.verbosity = 3 # 输出提示信息
# ?sc.settings.verbosity
sc.logging.print_header()
sc.settings.set_figure_params(dpi=80, facecolor='white')# 设置输出图像格式
results_file = 'write/pbmc3k.h5ad' # 存储分析结果
scanpy==1.6.0 anndata==0.7.5 umap==0.4.6 numpy==1.19.2 scipy==1.4.1 pandas==1.1.3 scikit-learn==0.23.2 statsmodels==0.12.0
这里的读取文件的方式和R语言构造seurat对象基本一样 (按照官网分类有12中读取方式)
下面主要介绍两种方法
第一种方法是,文件下面要有3个初始文件包括:
- barcord
- genes
- matrix
然后使用输sc.read_10_mtx读取
第二种方法是直接构建AnnData对象
然后分别的将表达矩阵,细胞信息,基因信息读取,代码如下
# 这个是第二种方法
#adata = sc.AnnData(counts.values, obs=cellinfo, var=geneinfo)
#adata.obs_names = cellinfo.Cell
#adata.var_names = geneinfo.Gene
#sc.pp.filter_genes(adata, min_counts=1)
#adata
# 这个是第一种读取方法
adata = sc.read_10x_mtx(
'./filtered_gene_bc_matrices/hg19/', # the directory with the `.mtx` file
var_names='gene_symbols', # use gene symbols for the variable names (variables-axis index)
cache=True)
adata.var_names_make_unique()
adata
tips: pytho和R语言有点不同,通常情况下,行为样本, 列为特征
adata.obs.shape # 2700个细胞
adata.var.shape # 32738个基因
adata.to_df().shape # 2700*32738
adata.obs.head()
adata.var.head()
adata.to_df().iloc[0:5,0:5]
数据预处理
这里介绍一下scanpy中常用的组件
- pp: 数据预处理
- tl: 添加额外信息
- pl:可视化
统计基因在细胞中的占比并可视化
sc.pl.highest_expr_genes(adata, n_top=20) # 每一个基因在所有细胞中的平均表达量(这里计算了百分比含量)
sc.pp.filter_cells(adata, min_genes=200) # 每一个细胞至少表达200个基因
sc.pp.filter_genes(adata, min_cells=3) # 每一个基因至少在3个细胞中表达
normalizing counts per cell
finished (0:00:00)
过滤线粒体DNA
adata.var['mt'] = adata.var_names.str.startswith('MT-')
adata.var['mt'].head()
# 抽取带有MT的字符串
adata.var['mt'] = adata.var_names.str.startswith('MT-')
# 数据过滤
sc.pp.calculate_qc_metrics(adata, qc_vars=['mt'], percent_top=None, log1p=False, inplace=True)
# 过滤后可视化(官方文档真的骚到我头皮发麻)
sc.pl.violin(adata, ['n_genes_by_counts'],jitter=0.4)
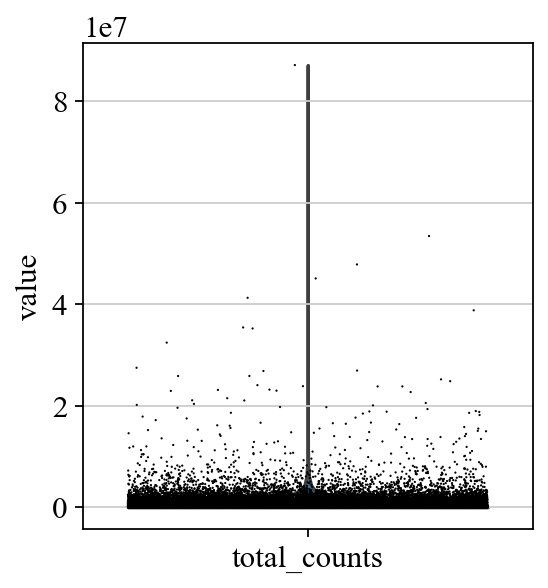
sc.pl.violin(adata, ['total_counts'],jitter=0.4)
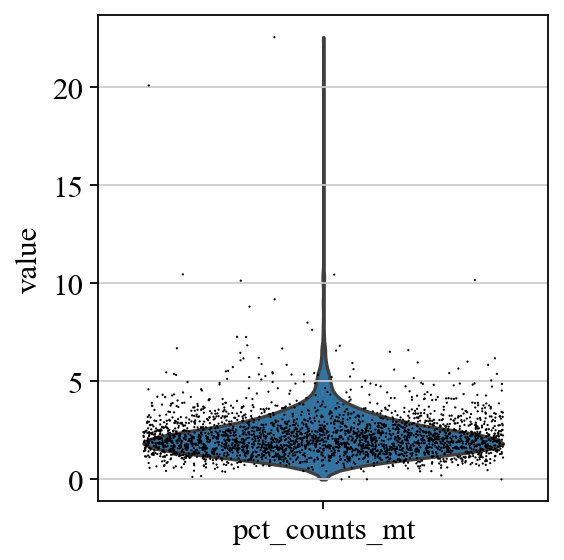
sc.pl.violin(adata, ['pct_counts_mt'],jitter=0.4)
sc.pl.scatter(adata, x='total_counts', y='pct_counts_mt')
sc.pl.scatter(adata, x='total_counts', y='n_genes_by_counts')
# 提取线粒体dna在5%以下
adata = adata[adata.obs.pct_counts_mt < 5, :]
# 提取基因不超过2500的细胞
adata = adata[adata.obs.n_genes_by_counts < 2500, :]
标准流程:
- log : NormalizeData
- 找特征 : FindVariableFeatures
- 标准化 : ScaleData
- pca : RunPCA
- 构建图 : FindNeighbors
- 聚类 : FindClusters
- tsne /umap : RunTSNE RunUMAP
- 差异基因 : FindAllMarkers / FindMarkers
sc.pp.normalize_total(adata, target_sum=1e4) # 不要和log顺序搞反了 ,这个是去文库的
sc.pp.log1p(adata)
sc.pp.highly_variable_genes(adata, min_mean=0.0125, max_mean=3, min_disp=0.5)
# 可视化
sc.pl.highly_variable_genes(adata)
# 保存一下原始数据
adata.raw = adata
# 提取高变基因
adata = adata[:, adata.var.highly_variable]
# 过滤掉没用的东西
sc.pp.regress_out(adata, ['total_counts', 'pct_counts_mt'])
# 中心化
sc.pp.scale(adata, max_value=10)
# pca
sc.tl.pca(adata, svd_solver='arpack')
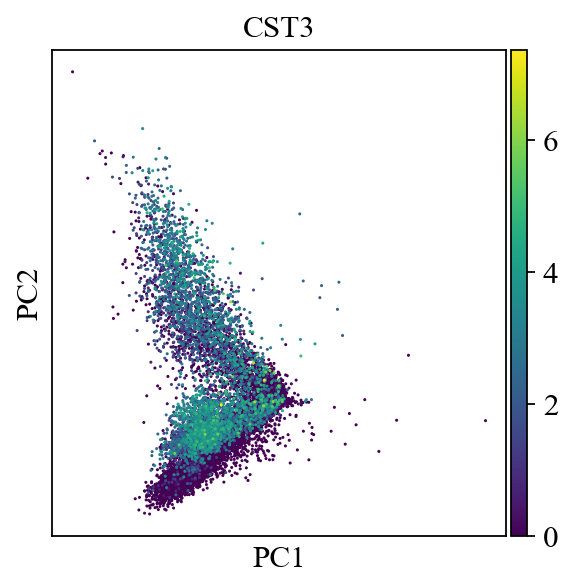
sc.pl.pca(adata, color='CST3')
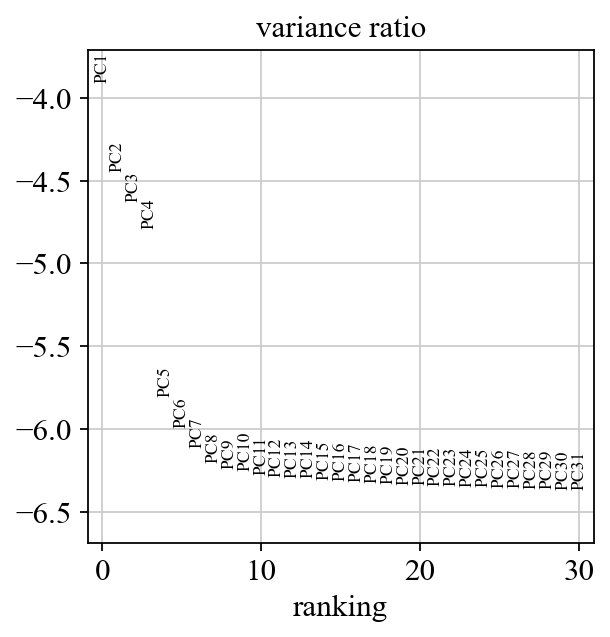
sc.pl.pca_variance_ratio(adata, log=True)
# 输出结果
adata.write(results_file)
# 构建图
sc.pp.neighbors(adata, n_neighbors=10, n_pcs=40)
sc.tl.umap(adata)
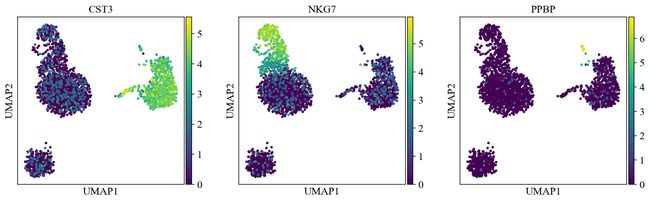
sc.pl.umap(adata, color=['CST3', 'NKG7', 'PPBP'])
sc.pl.umap(adata, color=['CST3', 'NKG7', 'PPBP'], use_raw=False)
sc.tl.tsne(adata)
sc.pl.tsne(adata, color=['CST3', 'NKG7', 'PPBP'])
sc.pl.tsne(adata, color=['CST3', 'NKG7', 'PPBP'], use_raw=False)
sc.pp.neighbors(adata, n_neighbors=10, n_pcs=40)
sc.tl.leiden(adata)
sc.pl.umap(adata, color=['leiden', 'CST3', 'NKG7'])
sc.pl.tsne(adata, color=['leiden', 'CST3', 'NKG7'])
# 保存结果
adata.write(results_file)
[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-5wQ4OHqI-1614138422679)(https://tva1.sinaimg.cn/large/0081Kckwly1glscmtf5q2j31hb0gj7cw.jpg)]
找差异基因
# 这里使用秩和检验
sc.tl.rank_genes_groups(adata, 'leiden', method='wilcoxon')
sc.pl.rank_genes_groups(adata, n_genes=25, sharey=False)
adata.write(results_file)
num = 2 # 通过这个控制marker基因的数量
marker_genes = list(set(np.array(pd.DataFrame(adata.uns['rank_genes_groups']['names']).head(num)).reshape(-1)))
len(marker_genes)
# 看一下每一个组的特征基因
adata = sc.read(results_file)
result = adata.uns['rank_genes_groups']
groups = result['names'].dtype.names
pd.DataFrame(
{group + '_' + key[:1]: result[key][group]
for group in groups for key in ['names', 'pvals']}).iloc[0:6,0:6]
# 比较组别间差异
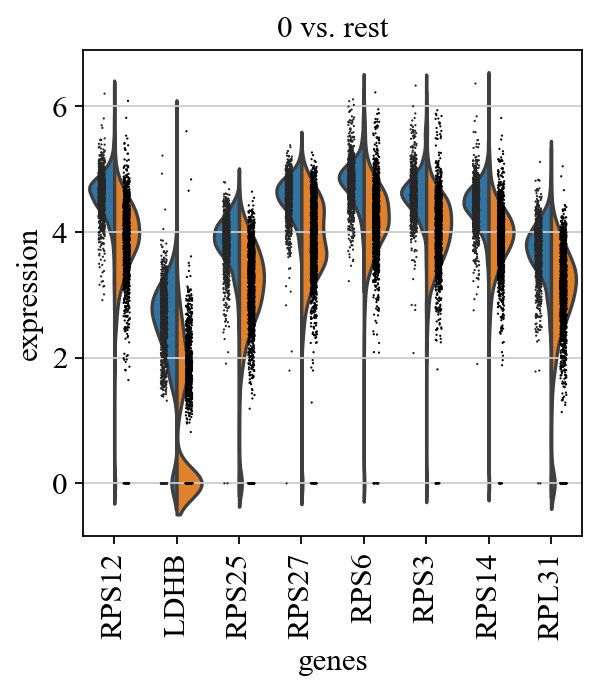
sc.tl.rank_genes_groups(adata, 'leiden', groups=['0'], reference='1', method='wilcoxon')
sc.pl.rank_genes_groups(adata, groups=['0'], n_genes=20)
sc.pl.rank_genes_groups_violin(adata, groups='0', n_genes=8)
# 这里需要重载一下结果,如果不重载的话结果会有差异的
adata = sc.read(results_file)
sc.pl.rank_genes_groups_violin(adata, groups='0', n_genes=8)
sc.pl.violin(adata, ['CST3', 'NKG7', 'PPBP'], groupby='leiden')
new_cluster_names = [
'CD4 T', 'CD14 Monocytes',
'B', 'CD8 T',
'NK', 'FCGR3A Monocytes',
'Dendritic', 'Megakaryocytes']
adata.rename_categories('leiden', new_cluster_names)
sc.pl.umap(adata, color='leiden', legend_loc='on data', title='', frameon=False, save='.pdf')
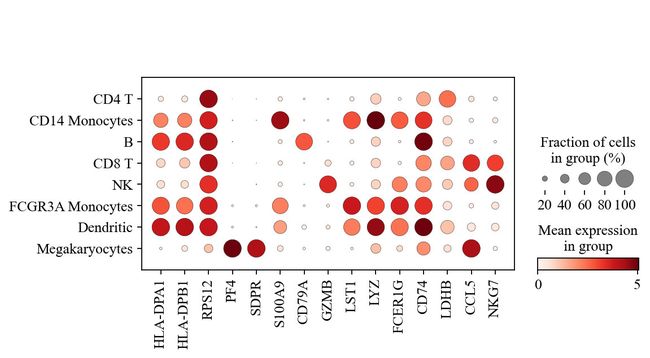
sc.pl.dotplot(adata, marker_genes, groupby='leiden');
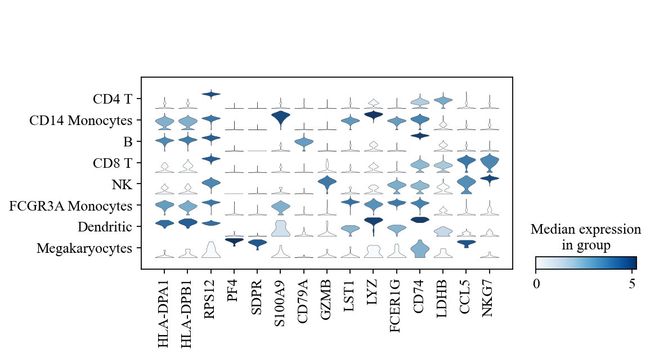
sc.pl.stacked_violin(adata, marker_genes, groupby='leiden', rotation=90);
adata.raw.to_adata().write('./write/pbmc3k_withoutX.h5ad')
WARNING: saving figure to file figures\umap.pdf
至此,标准流程构建完毕,然后试一下上次那篇cell的文章
import scanpy as sc
import os
import math
import itertools
import warnings
import numpy as np
import pandas as pd
import matplotlib.pyplot as plt
%matplotlib inline
%config InlineBackend.figure_format = 'svg'
warnings.filterwarnings("ignore")
plt.rc('font',family='Times New Roman')
my_colors = ["#1EB2A6","#ffc4a3","#e2979c","#F67575"]
sc.settings.verbosity = 3 # 输出提示信息
# ?sc.settings.verbosity
sc.logging.print_header()
sc.settings.set_figure_params(dpi=80, facecolor='white')# 设置输出图像格式
results_file = 'write/cell_reproduce.h5ad' # 存储分析结果
os.chdir('C:/Users/yuansh/OneDrive/课题/unfinishProgram/单细胞和自编码/scell_lung_adenocarcinoma-master/csv_files')
os.listdir()
这里的话介绍一下第二种数据导入的方法,原文中有2套数据,这里的话我就试一下第一套就行,后面的自行尝试
df = pd.read_csv('S01_datafinal.csv',index_col=0).T
cellinfo = pd.DataFrame(df.index,index=df.index,columns=['sample_index'])
geneinfo = pd.DataFrame(df.columns,index=df.columns,columns=['genes_index'])
adata = sc.AnnData(df, obs=cellinfo, var = geneinfo)
adata
接下来的步骤就是把上面的结果copy下来
sc.pl.highest_expr_genes(adata, n_top=20) # 每一个基因在所有细胞中的平均表达量(这里计算了百分比含量)
sc.pp.filter_cells(adata, min_genes=0) # 每一个细胞至少表达200个基因
sc.pp.filter_genes(adata, min_cells=0) # 每一个基因至少在3个细胞中表达
adata.var['mt'] = adata.var_names.str.startswith('MT-')
adata.var['mt'].head()
# 抽取带有MT的字符串
adata.var['mt'] = adata.var_names.str.startswith('MT-')
# 数据过滤
sc.pp.calculate_qc_metrics(adata, qc_vars=['mt'], percent_top=None, log1p=False, inplace=True)
# 这套数据是没有线粒体dna的
sc.pl.violin(adata, ['n_genes_by_counts'],jitter=0.4)
sc.pl.violin(adata, ['total_counts'],jitter=0.4)
sc.pl.violin(adata, ['pct_counts_mt'],jitter=0.4)
[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-KsWXv0rX-1614138422685)(https://tva1.sinaimg.cn/large/0081Kckwly1glscni9enzj30nc0lnacf.jpg)]
#再次强调这套数据没有线粒体dna
sc.pl.scatter(adata, x='total_counts', y='pct_counts_mt')
sc.pl.scatter(adata, x='total_counts', y='n_genes_by_counts')
# 提取线粒体dna在5%以下
adata = adata[adata.obs.pct_counts_mt < 5, :]
# 提取基因不超过2500的细胞
adata = adata[adata.obs.n_genes_by_counts < 2500, :]
sc.pp.normalize_total(adata, target_sum=1e4) # 不要和log顺序搞反了 ,这个是去文库的
sc.pp.log1p(adata)
sc.pp.highly_variable_genes(adata, min_mean=0.0125, max_mean=3, min_disp=0.5)
# 可视化
sc.pl.highly_variable_genes(adata)
# 保存一下原始数据
adata.raw = adata
[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-oUNLNImo-1614138422688)(https://tva1.sinaimg.cn/large/0081Kckwly1glsco1yoy3j30uz0guwgo.jpg)]
# 提取高变基因
adata = adata[:, adata.var.highly_variable]
# 过滤掉没用的东西
sc.pp.regress_out(adata, ['total_counts', 'pct_counts_mt'])
# 中心化
sc.pp.scale(adata, max_value=10)
# pca
sc.tl.pca(adata, svd_solver='arpack')
sc.pl.pca(adata, color='CST3')
sc.pl.pca_variance_ratio(adata, log=True)
adata.write(results_file) # 这里需要关闭一下百度网盘云同步
[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-XOFuNNkt-1614138422689)(https://tva1.sinaimg.cn/large/0081Kckwly1glsco74ippj30gb0hpaao.jpg)]
# 构建图
sc.pp.neighbors(adata, n_neighbors=10, n_pcs=40)
sc.tl.umap(adata)
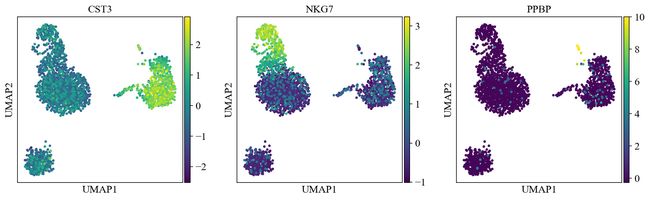
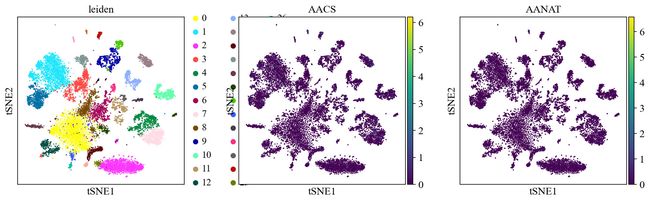
sc.pl.umap(adata, color=[ 'A4GALT', 'AACS', 'AANAT'])
sc.pl.umap(adata, color=[ 'A4GALT', 'AACS', 'AANAT'], use_raw=False)
sc.tl.tsne(adata)
sc.pl.tsne(adata, color=[ 'A4GALT', 'AACS', 'AANAT'])
sc.pl.tsne(adata, color=[ 'A4GALT', 'AACS', 'AANAT'], use_raw=False)
sc.pp.neighbors(adata, n_neighbors=10, n_pcs=40)
sc.tl.leiden(adata)
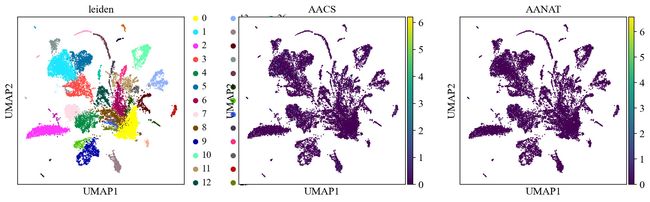
sc.pl.umap(adata, color=['leiden', 'AACS', 'AANAT'])
sc.pl.tsne(adata, color=['leiden', 'AACS', 'AANAT'])
# 保存结果
adata.write(results_file)
# 这里使用秩和检验
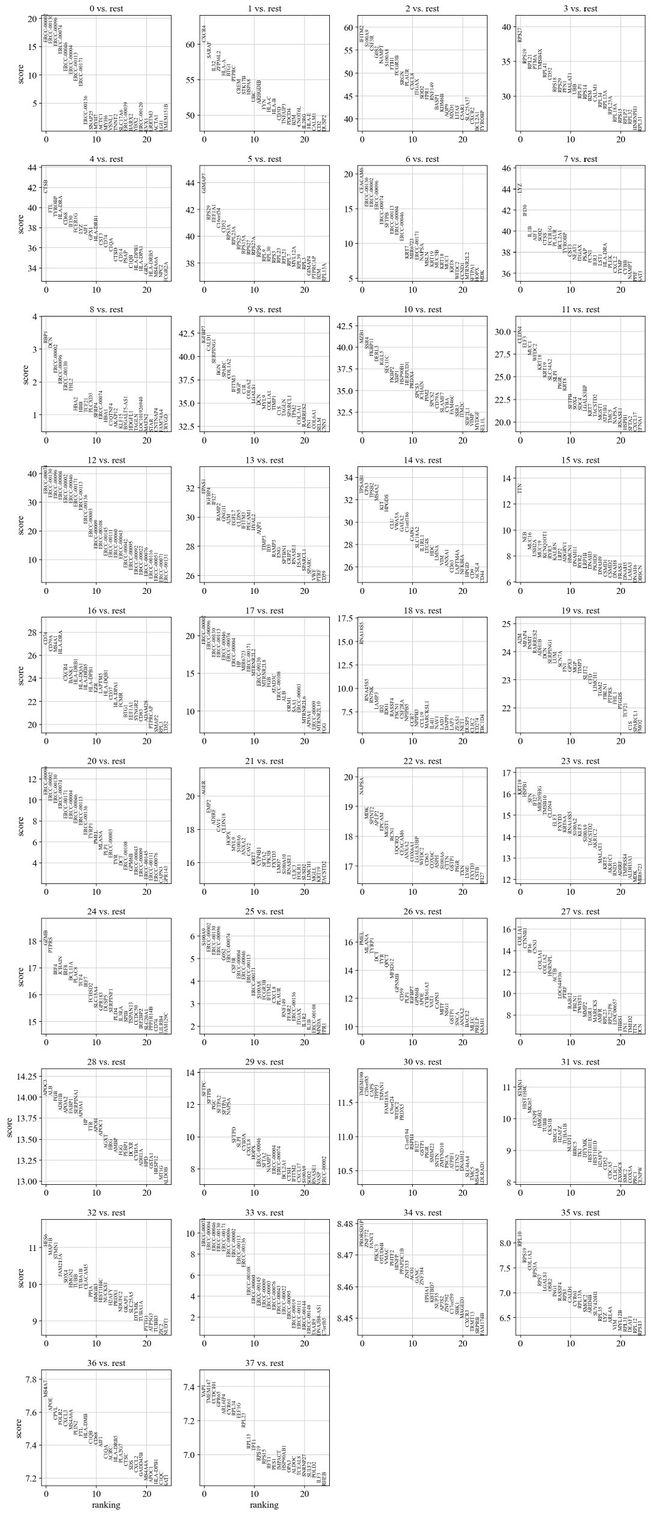
sc.tl.rank_genes_groups(adata, 'leiden', method='wilcoxon')
sc.pl.rank_genes_groups(adata, n_genes=25, sharey=False)
adata.write(results_file)
num = 2 # 通过这个控制marker基因的数量
marker_genes = list(set(np.array(pd.DataFrame(adata.uns['rank_genes_groups']['names']).head(num)).reshape(-1)))
len(marker_genes)
computing neighbors
using 'X_pca' with n_pcs = 40
finished: added to `.uns['neighbors']`
`.obsp['distances']`, distances for each pair of neighbors
`.obsp['connectivities']`, weighted adjacency matrix (0:00:02)
computing UMAP
finished: added
'X_umap', UMAP coordinates (adata.obsm) (0:00:06)
[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-aNHybOEv-1614138422690)(https://tva1.sinaimg.cn/large/0081Kckwly1glscoh4s05j31hq0gjk2t.jpg)]
[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-s0GDJaaO-1614138422690)(https://tva1.sinaimg.cn/large/0081Kckwly1glscok0km0j31hb0gjan8.jpg)]
[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-0Tce4rEo-1614138422691)(https://tva1.sinaimg.cn/large/0081Kckwly1glsconiccpj31hq0gjnai.jpg)]
# 看一下每一个组的特征基因
adata = sc.read(results_file)
result = adata.uns['rank_genes_groups']
groups = result['names'].dtype.names
pd.DataFrame(
{group + '_' + key[:1]: result[key][group]
for group in groups for key in ['names', 'pvals']}).iloc[0:6,0:6]
# 比较组别间差异
sc.tl.rank_genes_groups(adata, 'leiden', groups=['0'], reference='1', method='wilcoxon')
sc.pl.rank_genes_groups(adata, groups=['0'], n_genes=20)
[外链图片转存失败,源站可能有防盗链机制,建议将图片保存下来直接上传(img-eRqhgnzV-1614138422693)(https://tva1.sinaimg.cn/large/0081Kckwly1glscp2itwnj30h10hjjsc.jpg)]
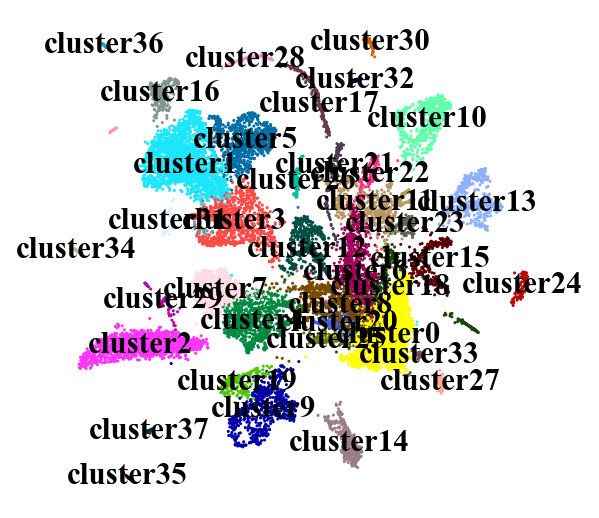
adata = sc.read(results_file)
new_cluster_names = ['cluster'+str(i) for i in range(0,38)]
adata.rename_categories('leiden', new_cluster_names)
sc.pl.umap(adata, color='leiden', legend_loc='on data', title='', frameon=False, save='.pdf')
sc.pl.dotplot(adata, marker_genes, groupby='leiden');
sc.pl.stacked_violin(adata, marker_genes, groupby='leiden', rotation=90);
如果您觉得我的文章对您有帮助,请点赞+关注,可以的话打个赏奖励一杯星巴克(~ ̄(OO) ̄)ブ
Best Regards,
Yuan.SH;
School of Basic Medical Sciences,
Fujian Medical University,
Fuzhou, Fujian, China.
please contact with me via the following ways:
(a) e-mail :[email protected]