TCGA样本分型
首先我们要定义所有样本的分型我们从TCGAbiolinks 中获取
library(TCGAbiolinks)
subtypes <- PanCancerAtlas_subtypes()
DT::datatable(subtypes,
filter = 'top',
options = list(scrollX = TRUE, keys = TRUE, pageLength = 5),
rownames = FALSE)
write.table(subtypes,file="sub.txt",quote=F,sep="\t")
这里获取的是TCGA所有样本的分型,需要再次筛选你需要的分型 ,这里以BRCA为例
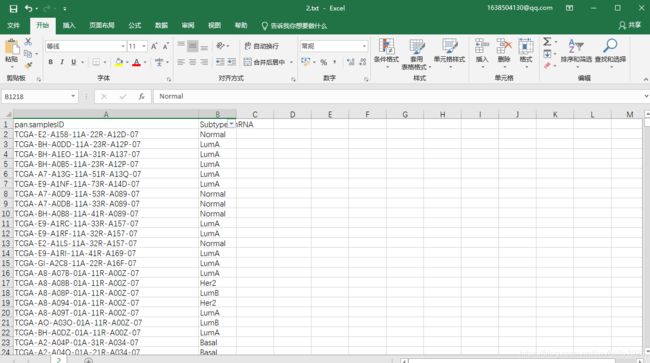
这里的文档要整理成这个样子
2 下面一步是为了进行数据的清洗
根据给的TCGA的样本我们进行划分,区分的97个case是有对照
首先清洗正常组的
rt <- read.table('2.txt',sep = '\t',header = T,check.names = F) ##导入的是上面表格的数据
rownames(rt) <- rt[,1]
tcga_data <- read.table('3.txt',sep = '\t',header = T,check.names = F)##导入99对那个数据
rownames(tcga_data) <- tcga_data[,1]
tcga_data <- tcga_data[,-1]
group_list=ifelse(as.numeric(substr(colnames(tcga_data),14,15)) < 10,'tumor','normal')##定义TCGA分组
tcga_normal <- tcga_data[, group_list == 'normal']##筛选正常组的数据
rt1=rt[match(colnames(tcga_normal),rt$pan.samplesID),] ##查看ID是否在样本分型的数据集中
rt2=rt1[rt1$Subtype_mRNA%in%c("Normal"),]###提取分型为正常的样本ID
tcga_normal_need=tcga_normal[,colnames(tcga_normal)%in%rt2$pan.samplesID]##整理样本分型为正常的样本
###> dim(tcga_normal_need)
###[1] 60483 87####97个中87的的癌旁是normal
2 接下来根据癌旁的结果选择对应癌做分析
首先我们看一下对应的癌是什么分型
tcga_tumor<- tcga_data[, group_list == 'tumor']
tcga_tumor_need <- tcga_tumor[,((substr(colnames(tcga_tumor),1,12))%in%(substr(rownames(rt2),1,12)))]###根据正常样本TCGA前12位来提取相同的向量的肿瘤的样本
tcga_need <- cbind(tcga_normal_need,tcga_tumor_need)##合并正常的与肿瘤
write.table(tcga_need,file = 'tcga_need.txt',sep = '\t',quote = F)
dim(tcga_need)
##> dim(tcga_need)
###[1] 60483 174
3 我们对肿瘤样本的分型进行定义
rt3=rt[match(colnames(tcga_tumor_need),rt$pan.samplesID),]##匹配ID并得到所有ID分型
LumA1 <-rt3[rt3$Subtype_mRNA%in%c("LumA"),]##LumA分型
LumB1 <- rt3[rt3$Subtype_mRNA%in%c("LumB"),]
Her21<- rt3[rt3$Subtype_mRNA%in%c("Her2"),]
Bascal1<- rt3[rt3$Subtype_mRNA%in%c("Basal"),]
Normal1<- rt3[rt3$Subtype_mRNA%in%c("Normal"),]
LumA=tcga_need[,colnames(tcga_need)%in%LumA1$pan.samplesID ]##LumA分型的数据
LumB=tcga_need[,colnames(tcga_need)%in%LumB1$pan.samplesID ]
Her2=tcga_need[,colnames(tcga_need)%in%Her21$pan.samplesID ]
Normal=tcga_need[,colnames(tcga_need)%in%Normal1$pan.samplesID ]
Bascal=tcga_need[,colnames(tcga_need)%in%Bascal1$pan.samplesID ]
data=cbind(LumA,LumB,Her2,Bascal,Normal)按照顺序进行分型的合并
这里展示一次分型对于肿瘤样本的分型接下来做PCA
library("FactoMineR")
library("factoextra")
library(tidyverse)
setwd("") #设置工作目录
data <- read.table("sample.txt", header=T, row.names=NULL,sep="\t")##数据导入
rownames_data <- make.names(data[,1],unique=T)##下面机组都是对数据的整理如果是接着做就不用了
data <- data[,-1,drop=F]
rownames(data) <- rownames_data
data <- data[rowSums(data) =!0,]###如果是上面图片的形式你可以从这一步开始,行不等于0
data <- data[apply(data, 1, var)!=0,] ##列的方差不为0
pca <- prcomp(t(data),scale = TRUE)##pca的核心
var_explained <- pca$sdev^2/sum(pca$sdev^2)
write.table(pca$rotation,file="PC.xls",quote=F,sep="\t") #输出特征向量
write.table(predict(pca),file="newTab.xls",quote=F,sep="\t") #输出新表
pca.sum=summary(pca)
write.table(pca.sum$importance,file="importance.xls",quote=F,sep="\t")#输出PC比重
pca$x %>%
as.data.frame %>%
ggplot(aes(x=PC1,y=PC2)) + geom_point(size=4) +
theme_bw(base_size=32) +
labs(x=paste0("PC1: ",round(var_explained[1]*100,1),"%"),
y=paste0("PC2: ",round(var_explained[2]*100,1),"%")) +
theme(legend.position="top")
pca$x %>%
as.data.frame %>%
rownames_to_column("continent_letter") %>%
separate(continent_letter,c("continent")) %>%
ggplot(aes(x=PC1,y=PC2)) + geom_point(aes(color=continent),size=4) +
theme_bw(base_size=32) +
labs(x=paste0("PC1: ",round(var_explained[1]*100,1),"%"),
y=paste0("PC2: ",round(var_explained[2]*100,1),"%")) +
theme(legend.position="top")
pca$x %>%
as.data.frame %>%
rownames_to_column("continent_typle") %>%
separate(continent_typle,c("continent", "typle")) %>%
ggplot(aes(x=PC1,y=PC2, label=typle, color=continent)) +
geom_label(aes(fill = continent), colour = "white", fontface = "bold")+
theme_bw(base_size=32) +
labs(x=paste0("PC1: ",round(var_explained[1]*100,1),"%"),
y=paste0("PC2: ",round(var_explained[2]*100,1),"%"))+
theme(legend.position="top")
pdf(file="pcaBarplot.pdf",width=15) #柱状图
barplot(pca.sum$importance[2,]*100,xlab="PC",ylab="percent",col="skyblue")
dev.off()
pdf(file="pcaPlot.pdf",width=15) #碎石图
plot(pca.sum$importance[2,]*100,type="o",col="red",xlab="PC",ylab="percent")
dev.off()
library(ggplot2)
##下面的分组是基于你前面data数据的顺序
group=c(rep("LumA",44),rep("LumB",17),rep("Her2",9),rep("Basal",14),rep("Normal",2)) #需要大家输入
pcaPredict=predict(pca)
PCA = data.frame(PCA1 = pcaPredict[,1], PCA2 = pcaPredict[,2],group=group)
PCA.mean=aggregate(PCA[,1:2],list(group=PCA$group),mean)
#定义椭圆
veganCovEllipse<-function (cov, center = c(0, 0), scale = 1, npoints = 100)
{
theta <- (0:npoints) * 2 * pi/npoints
Circle <- cbind(cos(theta), sin(theta))
t(center + scale * t(Circle %*% chol(cov)))
}
df_ell <- data.frame()
for(g in levels(PCA$group)){
df_ell <- rbind(df_ell, cbind(as.data.frame(with(PCA[PCA$group==g,],
veganCovEllipse(cov.wt(cbind(PCA1,PCA2),
wt=rep(1/length(PCA1),length(PCA1)))$cov,
center=c(mean(PCA1),mean(PCA2))))),group=g))
}
pdf(file="PCA2d.pdf")
ggplot(data = PCA, aes(PCA1, PCA2)) + geom_point(aes(color = group)) +
geom_path(data=df_ell, aes(x=PCA1, y=PCA2,colour=group), size=1, linetype=2)+
annotate("text",x=PCA.mean$PCA1,y=PCA.mean$PCA2,label=PCA.mean$group)+
theme_bw()+
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
dev.off()
library(TCGAbiolinks)
subtypes <- PanCancerAtlas_subtypes()
DT::datatable(subtypes,
filter = 'top',
options = list(scrollX = TRUE, keys = TRUE, pageLength = 5),
rownames = FALSE)
write.table(subtypes,file="sub.txt",quote=F,sep="\t")
rt <- read.table('2.txt',sep = '\t',header = T,check.names = F)
rownames(rt) <- rt[,1]
tcga_data <- read.table('3.txt',sep = '\t',header = T,check.names = F)
rownames(tcga_data) <- tcga_data[,1]
tcga_data <- tcga_data[,-1]
group_list=ifelse(as.numeric(substr(colnames(tcga_data),14,15)) < 10,'tumor','normal')
tcga_normal <- tcga_data[, group_list == 'normal']
rt1=rt[match(colnames(tcga_normal),rt$pan.samplesID),]
rt2=rt1[rt1$Subtype_mRNA%in%c("Normal"),]
tcga_normal_need=tcga_normal[,colnames(tcga_normal)%in%rt2$pan.samplesID]
dim(tcga_normal_need)
tcga_tumor<- tcga_data[, group_list == 'tumor']
tcga_tumor_need <- tcga_tumor[,((substr(colnames(tcga_tumor),1,12))%in%(substr(rownames(rt2),1,12)))]
tcga_need <- cbind(tcga_normal_need,tcga_tumor_need)
write.table(tcga_need,file = 'tcga_need.txt',sep = '\t',quote = F)
dim(tcga_need)
rt3=rt[match(colnames(tcga_tumor_need),rt$pan.samplesID),]
LumA1 <-rt3[rt3$Subtype_mRNA%in%c("LumA"),]
LumB1 <- rt3[rt3$Subtype_mRNA%in%c("LumB"),]
Her21<- rt3[rt3$Subtype_mRNA%in%c("Her2"),]
Bascal1<- rt3[rt3$Subtype_mRNA%in%c("Basal"),]
Normal1<- rt3[rt3$Subtype_mRNA%in%c("Normal"),]
LumA=tcga_need[,colnames(tcga_need)%in%LumA1$pan.samplesID ]
LumB=tcga_need[,colnames(tcga_need)%in%LumB1$pan.samplesID ]
Her2=tcga_need[,colnames(tcga_need)%in%Her21$pan.samplesID ]
Normal=tcga_need[,colnames(tcga_need)%in%Normal1$pan.samplesID ]
Bascal=tcga_need[,colnames(tcga_need)%in%Bascal1$pan.samplesID ]
data=cbind(LumA,LumB,Her2,Bascal,Normal)
data <- data[rowSums(data)>0,]
data <- data[apply(data, 1, var)!=0,]
pca <- prcomp(t(data),scale = TRUE)
var_explained <- pca$sdev^2/sum(pca$sdev^2)
write.table(pca$rotation,file="PC.xls",quote=F,sep="\t") #输出特征向量
write.table(predict(pca),file="newTab.xls",quote=F,sep="\t") #输出新表
pca.sum=summary(pca)
write.table(pca.sum$importance,file="importance.xls",quote=F,sep="\t")#输出PC比重
pca$x %>%
as.data.frame %>%
ggplot(aes(x=PC1,y=PC2)) + geom_point(size=4) +
theme_bw(base_size=32) +
labs(x=paste0("PC1: ",round(var_explained[1]*100,1),"%"),
y=paste0("PC2: ",round(var_explained[2]*100,1),"%")) +
theme(legend.position="top")
pca$x %>%
as.data.frame %>%
rownames_to_column("continent_letter") %>%
separate(continent_letter,c("continent")) %>%
ggplot(aes(x=PC1,y=PC2)) + geom_point(aes(color=continent),size=4) +
theme_bw(base_size=32) +
labs(x=paste0("PC1: ",round(var_explained[1]*100,1),"%"),
y=paste0("PC2: ",round(var_explained[2]*100,1),"%")) +
theme(legend.position="top")
pca$x %>%
as.data.frame %>%
rownames_to_column("continent_typle") %>%
separate(continent_typle,c("continent", "typle")) %>%
ggplot(aes(x=PC1,y=PC2, label=typle, color=continent)) +
geom_label(aes(fill = continent), colour = "white", fontface = "bold")+
theme_bw(base_size=32) +
labs(x=paste0("PC1: ",round(var_explained[1]*100,1),"%"),
y=paste0("PC2: ",round(var_explained[2]*100,1),"%"))+
theme(legend.position="top")
pdf(file="pcaBarplot.pdf",width=15) #柱状图
barplot(pca.sum$importance[2,]*100,xlab="PC",ylab="percent",col="skyblue")
dev.off()
pdf(file="pcaPlot.pdf",width=15) #碎石图
plot(pca.sum$importance[2,]*100,type="o",col="red",xlab="PC",ylab="percent")
dev.off()
library(ggplot2)
group=c(rep("LumA",44),rep("LumB",17),rep("Her2",9),rep("Basal",14),rep("Normal",2))
pcaPredict=predict(pca)
PCA = data.frame(PCA1 = pcaPredict[,1], PCA2 = pcaPredict[,2],group=group)
PCA.mean=aggregate(PCA[,1:2],list(group=PCA$group),mean)
#定义椭圆
veganCovEllipse<-function (cov, center = c(0, 0), scale = 1, npoints = 100)
{
theta <- (0:npoints) * 2 * pi/npoints
Circle <- cbind(cos(theta), sin(theta))
t(center + scale * t(Circle %*% chol(cov)))
}
df_ell <- data.frame()
for(g in levels(PCA$group)){
df_ell <- rbind(df_ell, cbind(as.data.frame(with(PCA[PCA$group==g,],
veganCovEllipse(cov.wt(cbind(PCA1,PCA2),
wt=rep(1/length(PCA1),length(PCA1)))$cov,
center=c(mean(PCA1),mean(PCA2))))),group=g))}
pdf(file="PCA2d.pdf")
ggplot(data = PCA, aes(PCA1, PCA2)) + geom_point(aes(color = group)) +
geom_path(data=df_ell, aes(x=PCA1, y=PCA2,colour=group), size=1, linetype=)+
annotate("text",x=PCA.mean$PCA1,y=PCA.mean$PCA2,label=PCA.mean$group)+
theme_bw()+
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
dev.off()
pdf(file="PCA2d.pdf")
ggplot(data = PCA, aes(PCA1, PCA2)) + geom_point(aes(color = group)) +
geom_path(data=df_ell, aes(x=PCA1, y=PCA2,colour=group), size=1, linetype=2)+
annotate("text",x=PCA.mean$PCA1,y=PCA.mean$PCA2,label=PCA.mean$group)+
theme_bw()+
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank())
dev.off()
