TPE原理总结
hyperopt - TPE
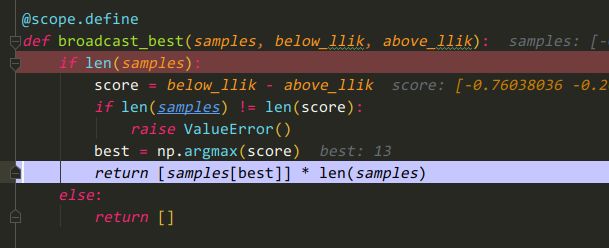
在hyperopt/tpe.py:935 处打一断点
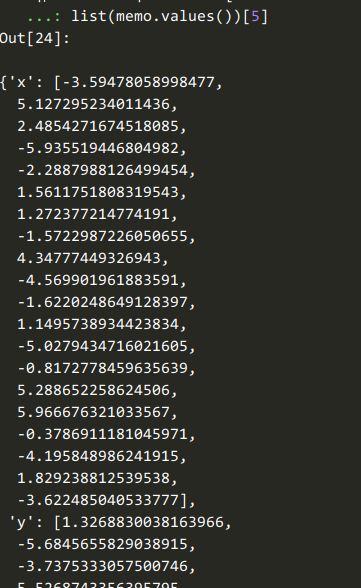
5:超参的取值
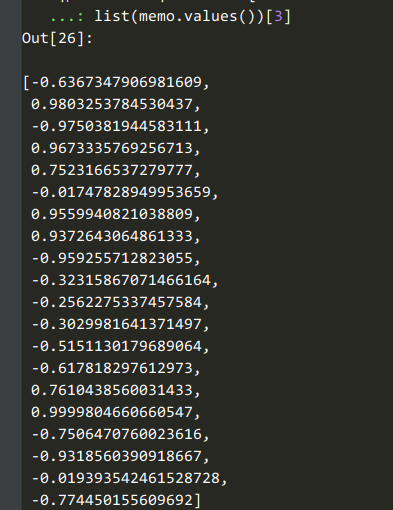
3:loss的取值
{'GMM1', 'GMM1_lpdf', 'len', 'dict', 'literal', 'broadcast_best', 'getitem', 'sub', 'array_union', 'mul', 'add',
'ap_split_trials', 'adaptive_parzen_normal', 'pos_args'}
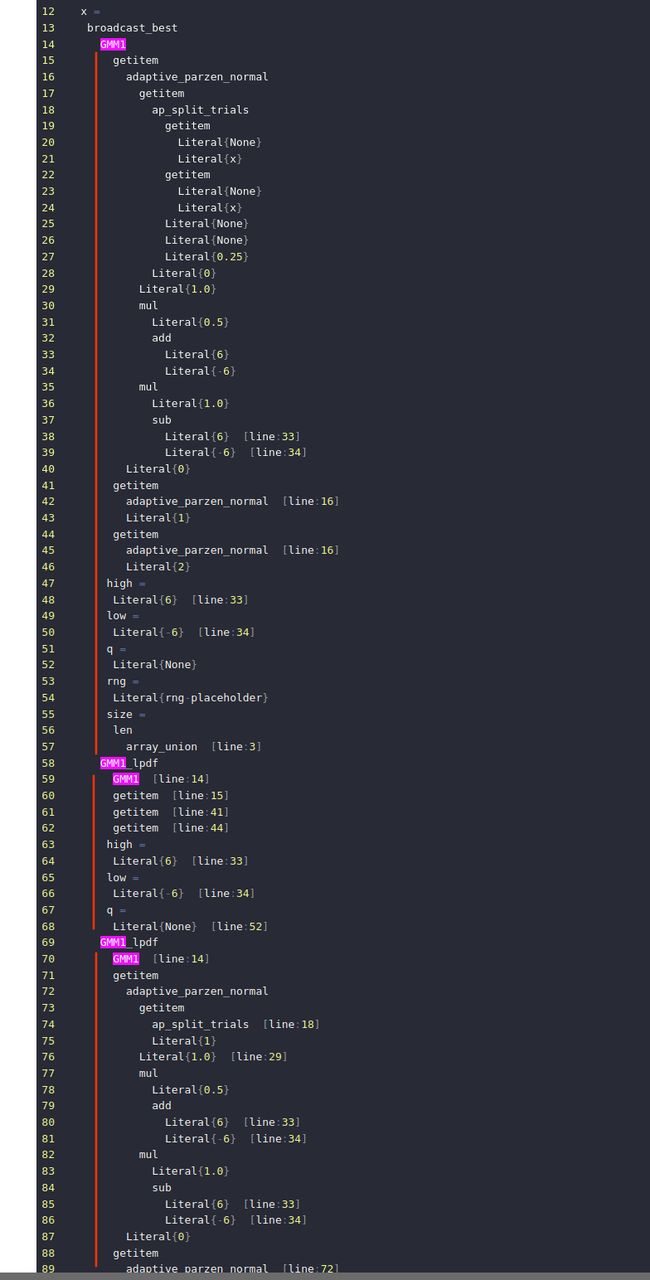
重点看一下hyperopt.tpe.build_posterior
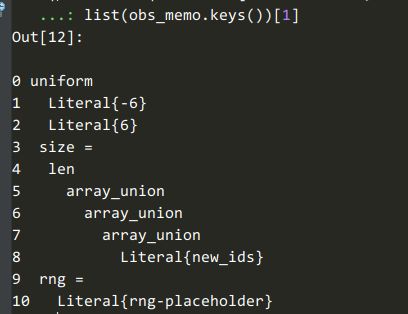
obs_memo 的 keys
采样函数: hyperopt/tpe.py:476
@adaptive_parzen_sampler("uniform")
def ap_uniform_sampler(obs, prior_weight, low, high, size=(), rng=None):
prior_mu = 0.5 * (high + low)
prior_sigma = 1.0 * (high - low)
weights, mus, sigmas = scope.adaptive_parzen_normal(
obs, prior_weight, prior_mu, prior_sigma
)
return scope.GMM1(
weights, mus, sigmas, low=low, high=high, q=None, size=size, rng=rng
)
@implicit_stochastic
@scope.define
def GMM1(weights, mus, sigmas, low=None, high=None, q=None, rng=None, size=()):
"""Sample from truncated 1-D Gaussian Mixture Model"""
weights, mus, sigmas = list(map(np.asarray, (weights, mus, sigmas)))
assert len(weights) == len(mus) == len(sigmas)
n_samples = int(np.prod(size))
# n_components = len(weights)
if low is None and high is None:
# -- draw from a standard GMM
active = np.argmax(rng.multinomial(1, weights, (n_samples,)), axis=1)
samples = rng.normal(loc=mus[active], scale=sigmas[active])
else:
# -- draw from truncated components, handling one-sided truncation
low = float(low) if low is not None else -float("Inf")
high = float(high) if high is not None else float("Inf")
if low >= high:
raise ValueError("low >= high", (low, high))
samples = []
while len(samples) < n_samples:
active = np.argmax(rng.multinomial(1, weights))
draw = rng.normal(loc=mus[active], scale=sigmas[active])
if low <= draw < high:
samples.append(draw)
samples = np.reshape(np.asarray(samples), size)
if q is None:
return samples
return np.round(old_div(samples, q)) * q
GMM1负责生成样本, GMM1_lpdf负责计算log likelihood
0 pos_args
1 dict
2 x =
3 array_union
4 array_union
5 array_union
6 Literal{new_ids}
7 y =
8 array_union
9 array_union
10 array_union [line:5]
11 dict
12 x =
13 broadcast_best
14 GMM1
15 getitem
16 adaptive_parzen_normal
17 getitem
18 ap_split_trials
19 getitem
20 Literal{None}
21 Literal{x}
22 getitem
23 Literal{None}
24 Literal{x}
25 Literal{None}
26 Literal{None}
27 Literal{0.25}
28 Literal{0}
29 Literal{1.0}
30 mul
31 Literal{0.5}
32 add
33 Literal{6}
34 Literal{-6}
35 mul
36 Literal{1.0}
37 sub
38 Literal{6} [line:33]
39 Literal{-6} [line:34]
40 Literal{0}
41 getitem
42 adaptive_parzen_normal [line:16]
43 Literal{1}
44 getitem
45 adaptive_parzen_normal [line:16]
46 Literal{2}
47 high =
48 Literal{6} [line:33]
49 low =
50 Literal{-6} [line:34]
51 q =
52 Literal{None}
53 rng =
54 Literal{rng-placeholder}
55 size =
56 len
57 array_union [line:3]
58 GMM1_lpdf
59 GMM1 [line:14]
60 getitem [line:15]
61 getitem [line:41]
62 getitem [line:44]
63 high =
64 Literal{6} [line:33]
65 low =
66 Literal{-6} [line:34]
67 q =
68 Literal{None} [line:52]
69 GMM1_lpdf
70 GMM1 [line:14]
71 getitem
72 adaptive_parzen_normal
73 getitem
74 ap_split_trials [line:18]
75 Literal{1}
76 Literal{1.0} [line:29]
77 mul
78 Literal{0.5}
79 add
80 Literal{6} [line:33]
81 Literal{-6} [line:34]
82 mul
83 Literal{1.0}
84 sub
85 Literal{6} [line:33]
86 Literal{-6} [line:34]
87 Literal{0}
88 getitem
89 adaptive_parzen_normal [line:72]
90 Literal{1}
91 getitem
92 adaptive_parzen_normal [line:72]
93 Literal{2}
94 high =
95 Literal{6} [line:33]
96 low =
97 Literal{-6} [line:34]
98 q =
99 Literal{None}
100 y =
101 broadcast_best
102 GMM1
103 getitem
104 adaptive_parzen_normal
105 getitem
106 ap_split_trials
107 getitem
108 Literal{None} [line:20]
109 Literal{y}
110 getitem
111 Literal{None} [line:23]
112 Literal{y}
113 Literal{None} [line:25]
114 Literal{None} [line:26]
115 Literal{0.25} [line:27]
116 Literal{0}
117 Literal{1.0} [line:29]
118 mul
119 Literal{0.5}
120 add
121 Literal{6}
122 Literal{-6}
123 mul
124 Literal{1.0}
125 sub
126 Literal{6} [line:121]
127 Literal{-6} [line:122]
128 Literal{0}
129 getitem
130 adaptive_parzen_normal [line:104]
131 Literal{1}
132 getitem
133 adaptive_parzen_normal [line:104]
134 Literal{2}
135 high =
136 Literal{6} [line:121]
137 low =
138 Literal{-6} [line:122]
139 q =
140 Literal{None}
141 rng =
142 Literal{rng-placeholder} [line:54]
143 size =
144 len
145 array_union [line:8]
146 GMM1_lpdf
147 GMM1 [line:102]
148 getitem [line:103]
149 getitem [line:129]
150 getitem [line:132]
151 high =
152 Literal{6} [line:121]
153 low =
154 Literal{-6} [line:122]
155 q =
156 Literal{None} [line:140]
157 GMM1_lpdf
158 GMM1 [line:102]
159 getitem
160 adaptive_parzen_normal
161 getitem
162 ap_split_trials [line:106]
163 Literal{1}
164 Literal{1.0} [line:29]
165 mul
166 Literal{0.5}
167 add
168 Literal{6} [line:121]
169 Literal{-6} [line:122]
170 mul
171 Literal{1.0}
172 sub
173 Literal{6} [line:121]
174 Literal{-6} [line:122]
175 Literal{0}
176 getitem
177 adaptive_parzen_normal [line:160]
178 Literal{1}
179 getitem
180 adaptive_parzen_normal [line:160]
181 Literal{2}
182 high =
183 Literal{6} [line:121]
184 low =
185 Literal{-6} [line:122]
186 q =
187 Literal{None}