.pth文件转.weight文件For YOLO
.pth文件转.weight文件
任务介绍
首先,.pth是pytorch框架训练模型的常见保存格式,.weight是darknet框架训练和加载模型的扩展名,实现将.pth转为.weight便可以将基于pytorch训练的模型在darknet框架里进行应用,比如作为预训练模型或直接进行检测。要做这件事,首先,咱得整明白下面这些东西:
- 怎么给参数从.pth文件中正确地拿出来,以及怎么给参数按照.weight文件需求的写进去
- 权重文件的存储规则:权重文件中哪部分是头文件,哪部分是网络参数,头文件都写了些啥;
- 网络参数的存储规则:网络中都有哪些模块有参数,一个模块中各个参数的存储顺序是啥,不同模块之间存储顺序是啥等等
前期查阅的资料
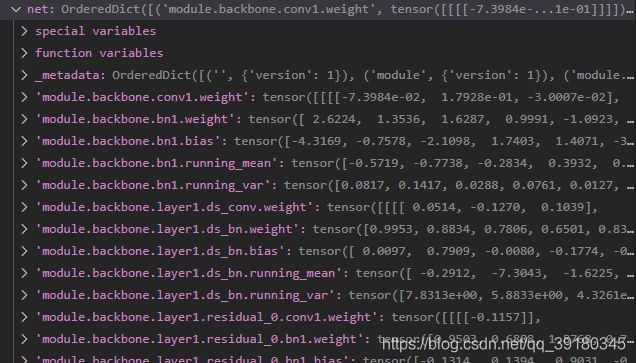
- torch.load可以解析.pth文件,得到参数存储的键值对,这样就可以直接获取到对应层的权重,随心所欲进行转换
net = torch.load(src_file,map_location=torch.device('cpu'))
- 一个讲了.weight文件的头文件应该咋写才能被darknet读取,链接
- 一个指明了转换的前进方向(.weight文件参数部分都是谁的参数,每个模块写的顺序是什么),链接

- 猜测darknet按照config文件进行权重读取,所以写.weight文件的时候应该和config保持一致
代码及注释
这里实现了对official_yolov3_weights_pytorch.pth的转换,这个权重是在pytorch框架中用YOLOv3算法训练的模型的权重,还特地torch.load了一下,确保参数是可以和darknet里yolov3的config文件对上的,这样转换之后的权重就可以很方便的在darknet中得到验证。
做这个实验就是为了验证现有方法对卷积核等权重导入的顺序(NCHW到底拉成一维向量是怎么拉的)是否正确,主要是也没找到明确的关于.weight文件中对于卷积核权重的存储顺序的说明,生怕现有的转换代码出现偏差,所以想着验证一下。验证的思路也比较蠢,就是想着找个训练的比较好的.pth文件,然后转换为.weight文件,然后在darknet框架中做个测试,如果检测结果不错,那证明当前的转换的代码没啥问题。
下面就贴上代码,这只实现了针对yolov3的权重转换,但是思路是一样的,改成resnet或其他网络结构只需要把load的顺序配合着darknet中的config来写就行。
明白这几点就可以:
1,torch.load()可以解析.pth中存储的参数键值对,还是按照顺序存储的,在一定程度上可以反映出网络结构
2,config中带有bn的conv,其参数写入.weight文件时的顺序:
‘bn1.bias’,‘bn1.weight’,‘bn1.running_mean’,‘bn1.running_var’,‘conv1.weight’
3,config中不带bn的conv,其参数写入.weight文件时的顺序:
‘conv.bias’,‘conv.weight’
4,对于卷积核的权重,其大小为NxCxHxW,从.pth中索引出conv_weight之后直接借助numpy的tofile()来实现拉成一维向量即可匹配.weight文件正确的卷积核参数存储顺序,像这样conv_weight.data.cpu().numpy().tofile(fp)
5,draknet是按照config文件写的顺序来导入权重的,route,shortcut这些层不影响导入顺序
import torch
import numpy as np
# list the path of the two kind of weight file below
src_file = '/disk2/pretrained_model/official_yolov3_weights_pytorch.pth'
dst_file = '/disk2/pretrained_model/yolov3.weight'
####################################################### structure of yolov3 ######################################################
# backbone part
backbone = ['module.backbone.',
'module.backbone.layer1.','module.backbone.layer1.residual', #1
'module.backbone.layer2.','module.backbone.layer2.residual', #2
'module.backbone.layer3.','module.backbone.layer3.residual', #8
'module.backbone.layer4.','module.backbone.layer4.residual', #8
'module.backbone.layer5.','module.backbone.layer5.residual' #4
]
num_of_residual = {'layer1':1,'layer2':2,'layer3':8,'layer4':8,'layer5':4}
ds_convbn = ['ds_bn.bias','ds_bn.weight','ds_bn.running_mean','ds_bn.running_var','ds_conv.weight']
convbn1 = ['bn1.bias','bn1.weight','bn1.running_mean','bn1.running_var','conv1.weight']
convbn2 = ['bn2.bias','bn2.weight','bn2.running_mean','bn2.running_var','conv2.weight']
# head part
embeddings = ['module.embedding0.','module.embedding1_cbl.','module.embedding1.','module.embedding2_cbl.','module.embedding2.']
convbn = ['bn.bias','bn.weight','bn.running_mean','bn.running_var','conv.weight']
conv = ['conv_out.bias','conv_out.weight']
####################################################### load the .pth file #######################################################
net = torch.load(src_file,map_location=torch.device('cpu'))
#################################################### write the .weight files #####################################################
# open a empty file and start to write
fp = open(dst_file, "wb")
# write head infomation into the file
header_info = np.array([0, 2, 0, 32013312, 0], dtype=np.int32)
header_info.tofile(fp)
# write the backbone part
for layer in backbone:
if layer.split('.')[-2] == 'backbone':
for i in convbn1:
content = net[layer+i]
content.data.cpu().numpy().tofile(fp)
if layer.split('.')[-2] == 'layer1':
if layer.split('.')[-1] =='':
# load the downsample part
for i in ds_convbn:
content = net[layer+i]
content.data.cpu().numpy().tofile(fp)
else:
# load the residual part
for j in range(num_of_residual['layer1']):
layer_new = layer+'_'+str(j)+'.'
for i in convbn1:
content = net[layer_new+i]
content.data.cpu().numpy().tofile(fp)
for i in convbn2:
content = net[layer_new+i]
content.data.cpu().numpy().tofile(fp)
if layer.split('.')[-2] == 'layer2':
if layer.split('.')[-1] =='':
# load the downsample part
for i in ds_convbn:
content = net[layer+i]
content.data.cpu().numpy().tofile(fp)
else:
# load the residual part
for j in range(num_of_residual['layer2']):
layer_new = layer+'_'+str(j)+'.'
for i in convbn1:
content = net[layer_new+i]
content.data.cpu().numpy().tofile(fp)
for i in convbn2:
content = net[layer_new+i]
content.data.cpu().numpy().tofile(fp)
if layer.split('.')[-2] == 'layer3':
if layer.split('.')[-1] =='':
# load the downsample part
for i in ds_convbn:
content = net[layer+i]
content.data.cpu().numpy().tofile(fp)
else:
# load the residual part
for j in range(num_of_residual['layer3']):
layer_new = layer+'_'+str(j)+'.'
for i in convbn1:
content = net[layer_new+i]
content.data.cpu().numpy().tofile(fp)
for i in convbn2:
content = net[layer_new+i]
content.data.cpu().numpy().tofile(fp)
if layer.split('.')[-2] == 'layer4':
if layer.split('.')[-1] =='':
# load the downsample part
for i in ds_convbn:
content = net[layer+i]
content.data.cpu().numpy().tofile(fp)
else:
# load the residual part
for j in range(num_of_residual['layer4']):
layer_new = layer+'_'+str(j)+'.'
for i in convbn1:
content = net[layer_new+i]
content.data.cpu().numpy().tofile(fp)
for i in convbn2:
content = net[layer_new+i]
content.data.cpu().numpy().tofile(fp)
if layer.split('.')[-2] == 'layer5':
if layer.split('.')[-1] =='':
# load the downsample part
for i in ds_convbn:
content = net[layer+i]
content.data.cpu().numpy().tofile(fp)
else:
# load the residual part
for j in range(num_of_residual['layer5']):
layer_new = layer+'_'+str(j)+'.'
for i in convbn1:
content = net[layer_new+i]
content.data.cpu().numpy().tofile(fp)
for i in convbn2:
content = net[layer_new+i]
content.data.cpu().numpy().tofile(fp)
# write the head part
for embedding in embeddings:
if embedding.split('_')[-1] == 'cbl.':
for i in convbn:
content = net[embedding+i]
content.data.cpu().numpy().tofile(fp)
else:
for j in range(6):
embedding_new = embedding+str(j)+'.'
for i in convbn:
content = net[embedding_new+i]
content.data.cpu().numpy().tofile(fp)
for i in conv:
content = net[embedding+i]
content.data.cpu().numpy().tofile(fp)
fp.close()
# finish !
把转换之后的权重使用darknet框架进行测试
./darknet detect cfg/yolov3.cfg pretrain_model/yolov3.weight data/dog.jpg
得到如下的检测结果
你看看!这狗多狗!这说明我们转换的权重是么得问题的~
目前只能说是针对模型来写转换的代码,而且因为.weight文件不存储网络结构,只能配合config文件加载权重,所以还不知道如果想导入的模块不连续或者不是从头开始该如何实现一次性导入,可能如果是想转换权重来做预训练可能还不太有这个需求
猜的不一定对,要是有更好的转换办法还麻烦评论区交流交流,互相学习呀~