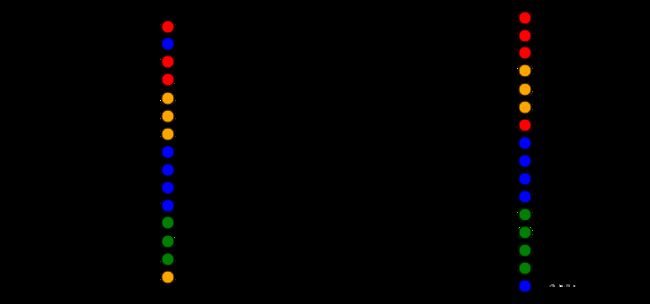Scikit-network-07:层次聚类Hierarchy
Hierarchy|层次聚类
Paris
介绍基于Paris算法的图层次聚类, Paris是基于linkage算聚类距离的。
from IPython.display import SVG
import numpy as np
from sknetwork.data import karate_club, painters, movie_actor
from sknetwork.hierarchy import Paris, cut_straight, dasgupta_score, tree_sampling_divergence
from sknetwork.visualization import svg_graph, svg_digraph, svg_bigraph, svg_dendrogram
图
graph = karate_club(metadata=True)
adjacency = graph.adjacency
position = graph.position
paris = Paris()
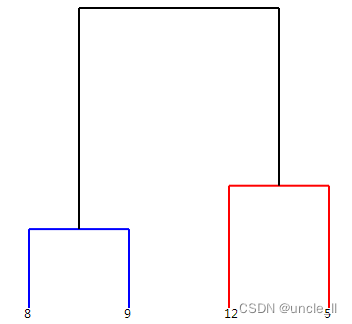
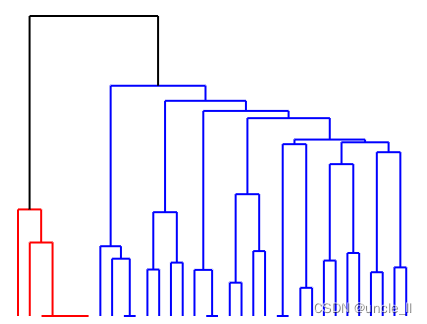
dendrogram = paris.fit_transform(adjacency) # 谱系结构
image = svg_dendrogram(dendrogram)
SVG(image)
# cuts of the dendrogram
labels = cut_straight(dendrogram)
print(labels)
# [1 1 1 1 1 1 1 1 0 1 1 1 1 1 0 0 1 1 0 1 0 1 0 0 0 0 0 0 0 0 0 0 0 0]
n_clusters = 4
labels, dendrogram_aggregate = cut_straight(dendrogram, n_clusters, return_dendrogram=True)
print(labels)
# [0 0 0 0 3 3 3 0 1 0 3 0 0 0 1 1 3 0 1 0 1 0 1 2 2 2 2 2 2 2 1 2 1 1]
_, counts = np.unique(labels, return_counts=True)
print(_, counts)
[0 1 2 3] [12 9 8 5]
# aggregate dendrogram
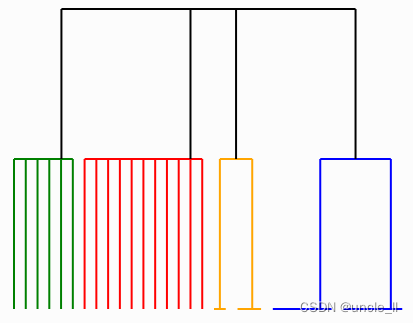
image = svg_dendrogram(dendrogram_aggregate, names=counts, rotate_names=False)
SVG(image)
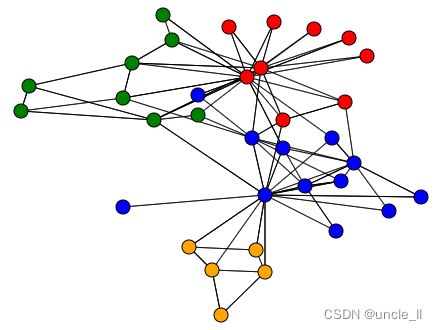
# corresponding clustering
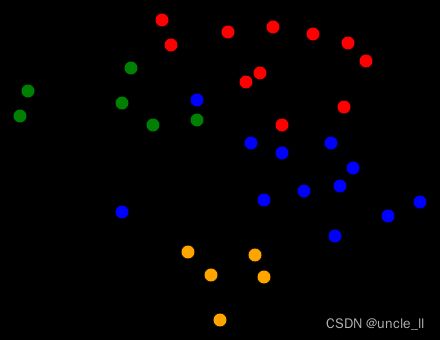
image = svg_graph(adjacency, position, labels=labels)
SVG(image)
有向图
graph = painters(metadata=True)
adjacency = graph.adjacency
position = graph.position
names = graph.names
# hierarchical clustering
paris = Paris()
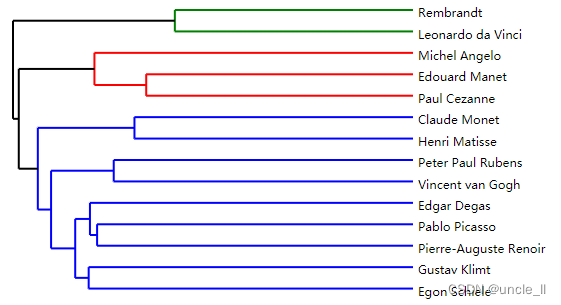
dendrogram = paris.fit_transform(adjacency)
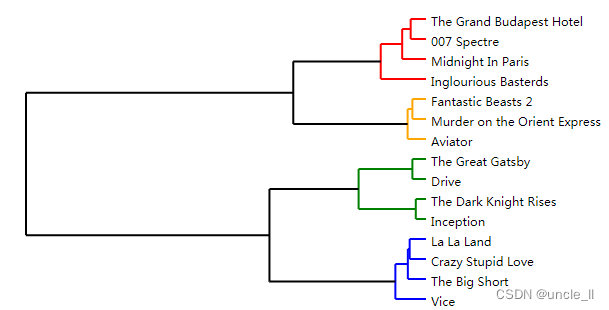
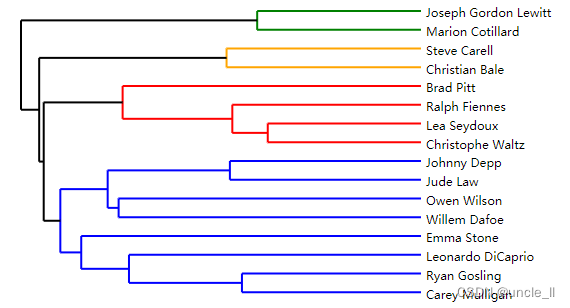
image = svg_dendrogram(dendrogram, names, n_clusters=3, rotate=True)
SVG(image)
# cut with 3 clusters
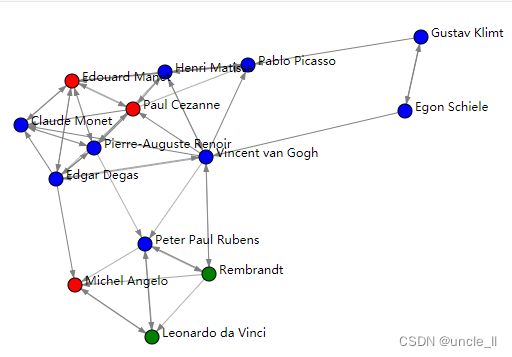
labels = cut_straight(dendrogram, n_clusters = 3)
print(labels)
# [0 0 1 0 1 1 2 0 0 1 0 0 0 2]
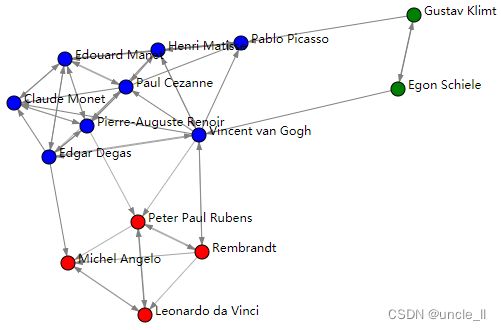
image = svg_digraph(adjacency, position, names=names, labels=labels)
SVG(image)
# metrics
dasgupta_score(adjacency, dendrogram)
# 0.5842857142857143
二部图
graph = movie_actor(metadata=True)
biadjacency = graph.biadjacency
names_row = graph.names_row
names_col = graph.names_col
# hierarchical clustering
paris = Paris()
paris.fit(biadjacency)
dendrogram_row = paris.dendrogram_row_
dendrogram_col = paris.dendrogram_col_
dendrogram_full = paris.dendrogram_full_
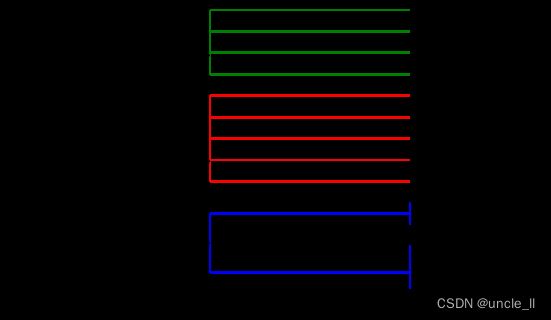
image = svg_dendrogram(dendrogram_row, names_row, n_clusters=4, rotate=True)
SVG(image)
image = svg_dendrogram(dendrogram_col, names_col, n_clusters=4, rotate=True)
SVG(image)
# cuts
labels = cut_straight(dendrogram_full, n_clusters=4)
n_row = biadjacency.shape[0]
labels_row = labels[:n_row]
labels_col = labels[n_row:]
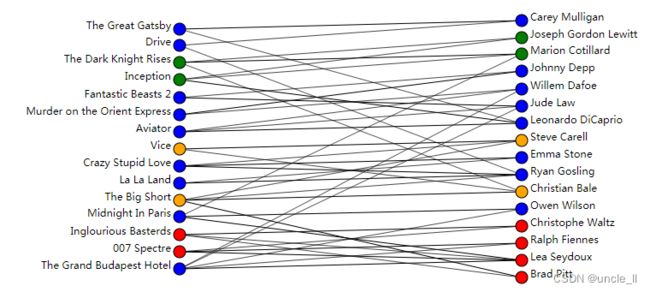
image = svg_bigraph(biadjacency, names_row, names_col, labels_row, labels_col)
SVG(image)
Ward
介绍基于Ward算法的图层次聚类, Ward是基于离差平方和ESS算聚类距离的。
from IPython.display import SVG
import numpy as np
from sknetwork.data import karate_club, painters, movie_actor
from sknetwork.embedding import Spectral
from sknetwork.hierarchy import Ward, cut_straight, dasgupta_score, tree_sampling_divergence
from sknetwork.visualization import svg_graph, svg_digraph, svg_bigraph, svg_dendrogram
图
graph = karate_club(metadata=True)
adjacency = graph.adjacency
position = graph.position
# hierachical clustering
ward = Ward()
dendrogram = ward.fit_transform(adjacency)
image = svg_dendrogram(dendrogram)
SVG(image)
# cuts
labels = cut_straight(dendrogram)
print(labels)
# [0 0 0 0 0 0 0 0 0 0 0 0 0 0 1 1 0 0 1 0 1 0 1 0 0 0 0 0 0 0 0 0 1 1]
n_cluster = 4
labels, dendrogram_aggregate = cut_straight(dendrogram, n_cluster, return_dendrogram=True)
print(labels)
# [0 2 0 3 0 0 0 3 0 0 0 0 3 3 1 1 0 2 1 2 1 2 1 0 0 0 0 0 0 0 0 0 1 1]
_, counts = np.unique(labels, return_counts=True)
# aggragate dendrogram
image = svg_dendrogram(dendrogram_aggregate, names=counts, rotate_names=False)
SVG(image)
# clustering
image = svg_graph(adjacency, position, labels=labels)
SVG(image)
# metrics
dasgupta_score(adjacency, dendrogram)
# 0.5082956259426847
# other embedding
ward = Ward(embedding_method=Spectral(4))
有向图
graph = painters(metadata=True)
adjacency = graph.adjacency
position = graph.position
names = graph.names
# hierarchical clustering
ward = Ward()
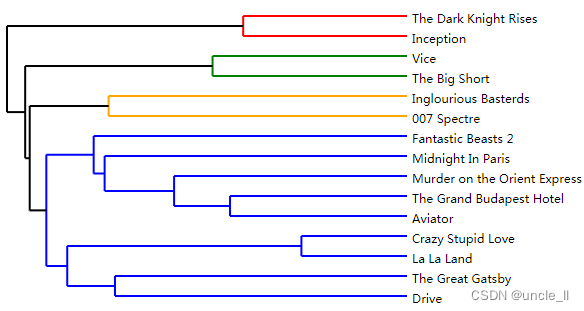
dendrogram = ward.fit_transform(adjacency)
image = svg_dendrogram(dendrogram, names, n_clusters=3, rotate=True)
SVG(image)
# cut with 3 clusters
labels = cut_straight(dendrogram, n_clusters=3)
print(labels)
# [0 0 1 1 0 2 0 0 0 2 0 1 0 0]
image = svg_digraph(adjacency, position, names=names, labels=labels)
SVG(image)
# metrics
dasgupta_score(adjacency, dendrogram)
# 0.31285714285714294
二部图
graph = movie_actor(metadata=True)
biadjacency = graph.biadjacency
names_row = graph.names_row
names_col = graph.names_col
# hierarchical clustering
ward = Ward(co_cluster=True)
ward.fit(biadjacency)
dendrogram_row = ward.dendrogram_row_
dendrogram_col = ward.dendrogram_col_
dendrogram_full = ward.dendrogram_full_
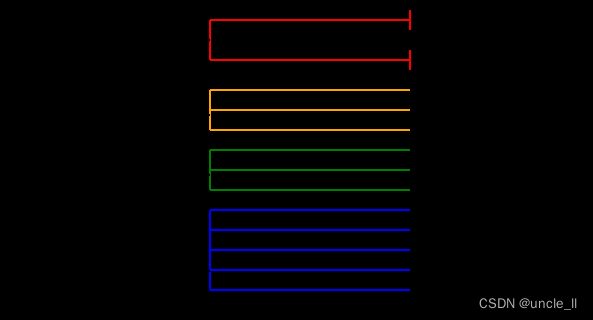
image = svg_dendrogram(dendrogram_row, names_row, n_clusters=4, rotate=True)
SVG(image)
image = svg_dendrogram(dendrogram_col, names_col, n_clusters=4, rotate=True)
SVG(image)
# cuts
labels = cut_straight(dendrogram_full, n_clusters = 4)
n_row = biadjacency.shape[0]
labels_row = labels[:n_row]
labels_col = labels[n_row:]
image = svg_bigraph(biadjacency, names_row, names_col, labels_row, labels_col)
SVG(image)
Louvain
介绍基于Louvain算法的图层次聚类, Louvain是基于模块度算聚类距离的。
from IPython.display import SVG
import numpy as np
from sknetwork.data import karate_club, painters, movie_actor
from sknetwork.hierarchy import LouvainHierarchy
from sknetwork.hierarchy import cut_straight, dasgupta_score, tree_sampling_divergence
from sknetwork.visualization import svg_graph, svg_digraph, svg_bigraph, svg_dendrogram
图
graph = karate_club(metadata=True)
adjacency = graph.adjacency
position = graph.position
# hierarchical clustering
louvain_hierarchy = LouvainHierarchy()
dendrogram = louvain_hierarchy.fit_transform(adjacency)
image = svg_dendrogram(dendrogram)
SVG(image)
# cuts
labels = cut_straight(dendrogram)
print(labels)
# [0 0 0 0 3 3 3 0 1 0 3 0 0 0 1 1 3 0 1 0 1 0 1 2 2 2 1 2 2 1 1 2 1 1]
labels, dendrogram_aggregate = cut_straight(dendrogram, n_clusters=4, return_dendrogram=True)
print(labels)
# [0 0 0 0 3 3 3 0 1 0 3 0 0 0 1 1 3 0 1 0 1 0 1 2 2 2 1 2 2 1 1 2 1 1]
_, counts = np.unique(labels, return_counts=True)
image = svg_dendrogram(dendrogram_aggregate, names=counts, rotate_names=False)
SVG(image)
image = svg_graph(adjacency, position, labels=labels)
SVG(image)
# metrics
dasgupta_score(adjacency, dendrogram)
# 0.6293363499245852
有向图
graph = painters(metadata=True)
adjacency = graph.adjacency
position = graph.position
names = graph.names
# hierarchical clustering
louvain_hierarchy = LouvainHierarchy()
dendrogram = louvain_hierarchy.fit_transform(adjacency)
image = svg_dendrogram(dendrogram, names, rotate=True)
SVG(image)
# cut with 3 clusters
labels = cut_straight(dendrogram, n_clusters=3)
print(labels)
# [1 0 2 0 2 2 1 0 1 2 1 0 0 1]
image = svg_digraph(adjacency, position, names=names, labels=labels)
SVG(image)
# metrics
dasgupta_score(adjacency, dendrogram)
# 0.53
二部图
graph = movie_actor(metadata=True)
biadjacency = graph.biadjacency
names_row = graph.names_row
names_col = graph.names_col
# hierarchical clustering
louvain_hierarchy = LouvainHierarchy()
louvain_hierarchy.fit(biadjacency)
dendrogram_row = louvain_hierarchy.dendrogram_row_
dendrogram_col = louvain_hierarchy.dendrogram_col_
dendrogram_full = louvain_hierarchy.dendrogram_full_
image = svg_dendrogram(dendrogram_row, names_row, n_clusters=4, rotate=True)
SVG(image)
image = svg_dendrogram(dendrogram_col, names_col, n_clusters=4, rotate=True)
SVG(image)
# cuts
labels = cut_straight(dendrogram_full, n_clusters=4)
n_row = biadjacency.shape[0]
labels_row = labels[:n_row]
labels_col = labels[n_row:]
image = svg_bigraph(biadjacency, names_row, names_col, labels_row, labels_col)
SVG(image)
参考
- https://scikit-network.readthedocs.io/en/latest/tutorials/hierarchy/louvain_recursion.html