【SimpleITK】CT数据的3D space归一化
- 导入依赖库
import SimpleITK as sitk
import matplotlib.pyplot as plt
- 示例数据
exaple_file = '1.nii.gz'
- 使用sampleITK读取数据,注意SimpleITK 加载数据是channel_first。
ds = sitk.ReadImage(exaple_file)
img_array = sitk.GetArrayFromImage(ds)
print(np.shape(img_array))
(150, 986, 512)
即 channel为150。
- 查看图像的原点Origin,大小Size,间距Spacing和方向Direction。
print(ds.GetOrigin())
print(ds.GetSize())
print(ds.GetSpacing())
print(ds.GetDirection())
(-454.462890625, 274.462890625, -1387.0)
(512, 986, 150)
(1.07421875, 1.07421875, 0.699999988079071)
(-1.0, 0.0, 0.0, 0.0, 1.0, 0.0, 0.0, 0.0, 1.0)
- 查看图像相关的纬度信息
print(ds.GetDimension())
print(ds.GetWidth())
print(ds.GetHeight())
print(ds.GetDepth())
3
512
986
150
- 体素类型查询
print(ds.GetPixelIDValue())
print(ds.GetPixelIDTypeAsString())
print(ds.GetNumberOfComponentsPerPixel())
2
16-bit signed integer
1
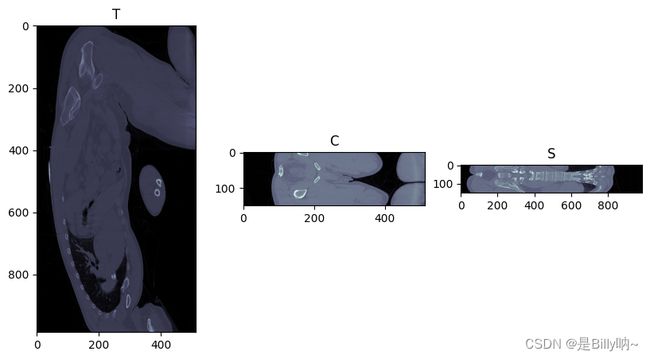
- 查看某一个横断面和冠状面
fig, (ax1, ax2, ax3) = plt.subplots(1, 3, figsize = (10, 5))
ax1.imshow(img_array[110,:,:], cmap=plt.cm.bone)
ax1.set_title('T')
ax2.imshow(img_array[:,110,:], cmap=plt.cm.bone)
ax2.set_title('C')
ax3.imshow(img_array[:,:,110], cmap=plt.cm.bone)
ax3.set_title('S')
- Resampling
def ImageResample(sitk_image, new_spacing = [1.0, 1.0, 1.0], is_label = False):
'''
sitk_image:
new_spacing: x,y,z
is_label: if True, using Interpolator `sitk.sitkNearestNeighbor`
'''
size = np.array(sitk_image.GetSize())
spacing = np.array(sitk_image.GetSpacing())
new_spacing = np.array(new_spacing)
new_size = size * spacing / new_spacing
new_spacing_refine = size * spacing / new_size
new_spacing_refine = [float(s) for s in new_spacing_refine]
new_size = [int(s) for s in new_size]
resample = sitk.ResampleImageFilter()
resample.SetOutputDirection(sitk_image.GetDirection())
resample.SetOutputOrigin(sitk_image.GetOrigin())
resample.SetSize(new_size)
resample.SetOutputSpacing(new_spacing_refine)
if is_label:
resample.SetInterpolator(sitk.sitkNearestNeighbor)
else:
#resample.SetInterpolator(sitk.sitkBSpline)
resample.SetInterpolator(sitk.sitkLinear)
newimage = resample.Execute(sitk_image)
return newimage
nor = ImageResample(ds)
print( nor.GetSize() )
(550, 1059, 104)