scanpy赋值问题
今天发现一个很奇怪的bug
import numpy as np
import pandas as pd
import anndata as ad
from scipy.sparse import csr_matrix
print(ad.__version__)
counts = csr_matrix(np.random.poisson(1, size=(100, 2000)), dtype=np.float32)
adata1 = ad.AnnData(counts)
print(adata1)
def f(adata):
adata = adata[:,0:1] #
print(adata.shape)
f(adata1)
print(adata1.shape)
结果如下

可以看到在函数中,这个adata的结果是变化了,但是并没有改变外部adata的值
import numpy as np
import pandas as pd
import anndata as ad
from scipy.sparse import csr_matrix
print(ad.__version__)
counts = csr_matrix(np.random.poisson(1, size=(100, 2000)), dtype=np.float32)
adata1 = ad.AnnData(counts)
print(adata1.X[0:2,0:10])
def f(adata):
adata = adata[:,0:1] #
print(adata.shape)
f(adata1)
print(adata1.shape)
print(adata1.X[0:2,0:10])
但是如果一开始我不在函数中操作,而是主程序中,这个结果
import numpy as np
import pandas as pd
import anndata as ad
from scipy.sparse import csr_matrix
print(ad.__version__)
counts = csr_matrix(np.random.poisson(1, size=(100, 2000)), dtype=np.float32)
adata1 = ad.AnnData(counts)
print(adata1.X.shape)
adata1 = adata1[:,0:1]
print(adata1.shape)
结果如下

这个现象只能解释为adata= adata1[:,0:1]是一个复制的行为,只不过同名了,所以adata的饮用变了,如果
adata2 = adata1[:,0:1],
可以想象,这个结果不会对adata1结果有影响
这仅仅是一个简简单单的例子,下面有一个更奇怪的测试
import scanpy as sc
adata= sc.read("/Users/yxk/Desktop/test_dataset/pbmc/pbmc.h5ad")
adata.obs["BATCH"] = adata.obs["batch"].copy()
adata.obs["label"]=adata.obs["celltype"].astype("category").cat.codes
n_classes= len(adata.obs["label"].value_counts())
print(adata)
adata1= adata[adata.obs["batch"]=="pbmc_3p"].copy()
adata2= adata[adata.obs["batch"]=="pbmc_5p"].copy()
#print(adata1.X)
#print(adata2.X)
## 如果用这种方式,我的结果是这样的
def preprocessNew(
adata_A_input,
):
'''
Performing preprocess for a pair of datasets.
To integrate multiple datasets, use function preprocess_multiple_anndata in utils.py
'''
adata_A = adata_A_input
print("Finding highly variable genes...")
#sc.pp.highly_variable_genes(adata_A, flavor='seurat_v3', n_top_genes=2000)
#hvg_A = adata_A.var[adata_A.var.highly_variable == True].sort_values(by="highly_variable_rank").index
print("Normalizing and scaling...")
sc.pp.normalize_total(adata_A, target_sum=1e4)
sc.pp.log1p(adata_A)
sc.pp.highly_variable_genes(adata_A,n_top_genes=2000)
hvg_A = list(adata1.var_names[adata1.var.highly_variable])
adata_A = adata_A[:, hvg_A]
sc.pp.scale(adata_A, max_value=10)
print(adata_A.X[0:1,0:100])
print(adata_A.X.shape)
# 为啥这些结果是这样的
preprocessNew(adata1)
print(adata1.X.shape)
 可以看到adata的结果是没有改变的,还是33694维,但是我在函数中,明明是选择了高变基因的
可以看到adata的结果是没有改变的,还是33694维,但是我在函数中,明明是选择了高变基因的
但是如果采用下面的代码
import scanpy as sc
adata= sc.read("/Users/yxk/Desktop/test_dataset/pbmc/pbmc.h5ad")
adata.obs["BATCH"] = adata.obs["batch"].copy()
adata.obs["label"]=adata.obs["celltype"].astype("category").cat.codes
n_classes= len(adata.obs["label"].value_counts())
print(adata)
adata1= adata[adata.obs["batch"]=="pbmc_3p"].copy()
adata2= adata[adata.obs["batch"]=="pbmc_5p"].copy()
#print(adata1.X)
#print(adata2.X)
def preprocessNew(
adata_A_input,
):
'''
Performing preprocess for a pair of datasets.
To integrate multiple datasets, use function preprocess_multiple_anndata in utils.py
'''
adata_A = adata_A_input
print("Finding highly variable genes...")
#sc.pp.highly_variable_genes(adata_A, flavor='seurat_v3', n_top_genes=2000)
#hvg_A = adata_A.var[adata_A.var.highly_variable == True].sort_values(by="highly_variable_rank").index
print("Normalizing and scaling...")
sc.pp.normalize_total(adata_A, target_sum=1e4)
sc.pp.log1p(adata_A)
sc.pp.highly_variable_genes(adata_A,n_top_genes=2000,subset=True)
#adata_A = adata_A[:, hvg_A]
sc.pp.scale(adata_A, max_value=10)
print(adata_A.X[0:1,0:100])
preprocessNew(adata1)
print(adata1.X.shape)
print(adata1.X[0:1,0:100])
## 但是线则这个问题为啥不是
结果如下
这里可以看到,我最终的adata1的维度是改变了,这里需要注意
这里使用
sc.pp.highly_variable_genes(adata1,n_top_genes=2000,subset=True),就是对adata的引用改动了,最终导致最开始的atata出现了变化,反正最好还是用scanpy的内置函数了,一旦在函数里赋值就要注意局部对象的问题
import scanpy as sc
adata= sc.read("/Users/yxk/Desktop/test_dataset/pbmc/pbmc.h5ad")
adata.obs["BATCH"] = adata.obs["batch"].copy()
adata.obs["label"]=adata.obs["celltype"].astype("category").cat.codes
n_classes= len(adata.obs["label"].value_counts())
print(adata)
adata1= adata[adata.obs["batch"]=="pbmc_3p"].copy()
#adata2= adata[adata.obs["batch"]=="pbmc_5p"].copy()
#print(adata1.X)
#print(adata2.X)
print("Normalizing and scaling...")
sc.pp.normalize_total(adata1, target_sum=1e4)
sc.pp.log1p(adata1)
sc.pp.highly_variable_genes(adata1,n_top_genes=2000,subset=True)
sc.pp.scale(adata1, max_value=10)
print(adata1.X[0:1,0:100])
print(adata1.X.shape)
print(adata1.X[0:1,0:100])
## 但是线则这个问题为啥不是
如果采用了preprocessNew的函数,那么本质上只对adata做了如下变化
import scanpy as sc
adata= sc.read("/Users/yxk/Desktop/test_dataset/pbmc/pbmc.h5ad")
adata.obs["BATCH"] = adata.obs["batch"].copy()
adata.obs["label"]=adata.obs["celltype"].astype("category").cat.codes
n_classes= len(adata.obs["label"].value_counts())
print(adata)
adata1= adata[adata.obs["batch"]=="pbmc_3p"].copy()
#adata2= adata[adata.obs["batch"]=="pbmc_5p"].copy()
#print(adata1.X)
#print(adata2.X)
## 如果用这种方式,我的结果是这样的
def preprocessNew(
adata_A_input,
):
'''
Performing preprocess for a pair of datasets.
To integrate multiple datasets, use function preprocess_multiple_anndata in utils.py
'''
adata_A = adata_A_input
print("Finding highly variable genes...")
#sc.pp.highly_variable_genes(adata_A, flavor='seurat_v3', n_top_genes=2000)
#hvg_A = adata_A.var[adata_A.var.highly_variable == True].sort_values(by="highly_variable_rank").index
print("Normalizing and scaling...")
sc.pp.normalize_total(adata_A, target_sum=1e4)
sc.pp.log1p(adata_A)
sc.pp.highly_variable_genes(adata_A,n_top_genes=2000)
hvg_A = list(adata1.var_names[adata1.var.highly_variable])
adata_A = adata_A[:, hvg_A]
sc.pp.scale(adata_A, max_value=10)
print(adata_A.X[0:1,0:100])
print(adata_A.X.shape)
# 为啥这些结果是这样的
preprocessNew(adata1)
print(adata1.X.shape)
print(adata1.X[0:1,0:100])
结果如下
reproduce result
import scanpy as sc
adata= sc.read("/Users/yxk/Desktop/test_dataset/pbmc/pbmc.h5ad")
adata.obs["BATCH"] = adata.obs["batch"].copy()
adata.obs["label"]=adata.obs["celltype"].astype("category").cat.codes
n_classes= len(adata.obs["label"].value_counts())
print(adata)
adata2= adata[adata.obs["batch"]=="pbmc_3p"].copy()
#print(adata1.X)
#print(adata2.X)
## 如果用这种方式,我的结果是这样的
print("Normalizing and scaling...")
sc.pp.normalize_total(adata2, target_sum=1e4)
sc.pp.log1p(adata2) # 真正对adata1只有这么多的操作
# 为啥这些结果是这样的
print(adata2.X.shape)
print(adata2.X[0:1,0:100])
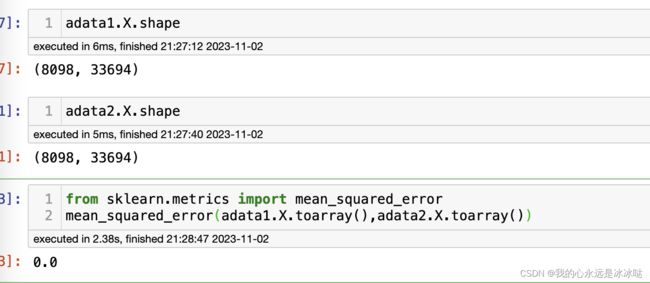
from sklearn.metrics import mean_squared_error
mean_squared_error(adata1.X.toarray(),adata2.X.toarray())