SVM
import numpy as np
from scipy import io as spio
from matplotlib import pyplot as plt
from sklearn import svm
def SVM():
'''data1——线性分类'''
data1 = spio.loadmat('data1.mat')
X = data1['X']
y = data1['y']
y = np.ravel(y)
plot_data(X, y)
model = svm.SVC(C=1.0, kernel='linear').fit(X, y)
plot_decisionBoundary(X, y, model)
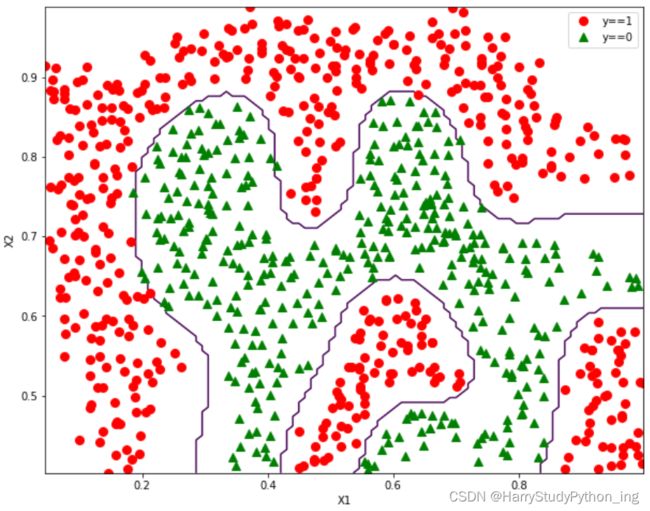
'''data2——非线性分类'''
data2 = spio.loadmat('data2.mat')
X = data2['X']
y = data2['y']
y = np.ravel(y)
plt = plot_data(X, y)
plt.show()
model = svm.SVC(gamma=100).fit(X, y)
plot_decisionBoundary(X, y, model, class_='notLinear')
def plot_data(X, y):
plt.figure(figsize=(10, 8))
pos = np.where(y == 1)
neg = np.where(y == 0)
p1, = plt.plot(np.ravel(X[pos, 0]), np.ravel(X[pos, 1]), 'ro', markersize=8)
p2, = plt.plot(np.ravel(X[neg, 0]), np.ravel(X[neg, 1]), 'g^', markersize=8)
plt.xlabel("X1")
plt.ylabel("X2")
plt.legend([p1, p2], ["y==1", "y==0"])
return plt
def plot_decisionBoundary(X, y, model, class_='linear'):
plt = plot_data(X, y)
if class_ == 'linear':
w = model.coef_
b = model.intercept_
xp = np.linspace(np.min(X[:, 0]), np.max(X[:, 0]), 100)
yp = -(w[0, 0] * xp + b) / w[0, 1]
plt.plot(xp, yp, 'b-', linewidth=2.0)
plt.show()
else:
x_1 = np.transpose(np.linspace(np.min(X[:, 0]), np.max(X[:, 0]), 100).reshape(1, -1))
x_2 = np.transpose(np.linspace(np.min(X[:, 1]), np.max(X[:, 1]), 100).reshape(1, -1))
X1, X2 = np.meshgrid(x_1, x_2)
vals = np.zeros(X1.shape)
for i in range(X1.shape[1]):
this_X = np.hstack((X1[:, i].reshape(-1, 1), X2[:, i].reshape(-1, 1)))
vals[:, i] = model.predict(this_X)
plt.contour(X1, X2, vals, [0, 1], color='blue')
plt.show()
if __name__ == "__main__":
SVM()