[论文精读]Large-scale Graph Representation Learning of Dynamic Brain Connectome with Transformers
论文网址:[2312.14939] Large-scale Graph Representation Learning of Dynamic Brain Connectome with Transformers (arxiv.org)
英文是纯手打的!论文原文的summarizing and paraphrasing。可能会出现难以避免的拼写错误和语法错误,若有发现欢迎评论指正!文章偏向于笔记,谨慎食用!
又见面了 Prof Kim
目录
1. 省流版
1.1. 心得
1.2. 论文框架图
2. 论文逐段精读
2.1. Abstract
2.2. Introduction
2.3. Main Contribution
2.3.1. Defining the Connectome Embedding
2.3.2. TENET: Temporal Neural Transformer
2.4. Experiments
2.4.1. Dataset and Experimental Setup
2.4.2. Comparative Experiment
2.4.3. Ablation Study
2.5. Conclusion
3. Reference List
1. 省流版
1.1. 心得
(1)都2023年了为啥开篇还在说“迄今为止的研究都低估了功能连接的时间动态”,好像还行吧我感觉读了几篇动态的了
(2)⭐五万!!!??个!?样本!!?
(3)⭐这是一个好问题,以至于我真的没有发现。FC随着时间改变有什么意义?为什么会改变?它应该是像血液循环或者呼吸那样有个周期吗,还是说有别的什么意思?为什么迄今为止没有人分析过这个呢?
(4)四页!
1.2. 论文框架图
2. 论文逐段精读
2.1. Abstract
①Previous works did not put dynamic character in a important position
②⭐The authors aim to learn dynamic functional connectivity (FC) by graph transformers (GTs)
③⭐They use over 50,000 rs-fMRI data from 3 datasets, which is the largest number in relevant researches so far
④Tasks: sex classification and age regression
2.2. Introduction
①When deal FC with GNN, the data might be over smoothing or over squashing. Besides, rich connectivity details can also be lost
②Node and edge embedding is still a challenge in GTs
③Large scale data can probably enhance the replicability
④⭐Exsting works do not reveal the nature of why FC will changes with time
2.3. Main Contribution
2.3.1. Defining the Connectome Embedding
①Get time series matrix ![]() in time
in time ![]()
②Encoding dynamic FC with original position, structure and time by sliding window and GRU based methods
③The edge weight:
where ![]() is a windowed matrix from
is a windowed matrix from ![]() window length and stride
window length and stride ![]() in time
in time ![]()
④Reduce identity matrix from ![]() to ensure there is no self-loop. Then combine it with another identity matrix:
to ensure there is no self-loop. Then combine it with another identity matrix:
![]()
put ![]() to two MLP layers then get a
to two MLP layers then get a ![]() dimensional input
dimensional input
⑤Time embedding ![]() is the GRU output (?). Combine the two will get a final connectome embedding:
is the GRU output (?). Combine the two will get a final connectome embedding:
![]()
⑥They effectively capture the one-hop connectivity information
⑦Connectome embedding method:
2.3.2. TENET: Temporal Neural Transformer
①The framework of TENET with two parts:
they firstly transform ![]() then excute
then excute ![]() as
as ![]() , where
, where ![]() is a self attended connectome feature vecture at time
is a self attended connectome feature vecture at time ![]() .
. ![]() aims to capture spatial features and
aims to capture spatial features and ![]() aims to learn temporal self-attention.
aims to learn temporal self-attention.
②The attention function:
where ![]() is the hidden dimension
is the hidden dimension
③MHSA means self-attention parallelly projected with the multiple number of heads
④Combine operator: ![]()
⑤Left MLP layer: ![]() and the first layer is
and the first layer is ![]()
⑥The detailed pseudo code of TENET(正文放在附录,但是只有一个附录我就把它提到正文了):
culminate vi.达到顶点;(以某种结果)告终;(在某一点)结束
2.4. Experiments
2.4.1. Dataset and Experimental Setup
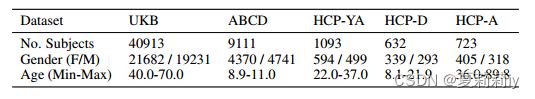
①Datasets: UK Biobank (UKB), Adolescent Brain Cognitive Development (ABCD) (they choose ABCD-HCP pipeline 2 for unpreprocessed data) and Human Connectome Project (HCP) with different preprocessing
②Atlas: Schaefer with 400 ROIs
③Layer of model: 4
④Number of hidden dimension in each layer: 1024
⑤Optimizer: Adam
⑥Search the best batch size by grid search in {2, 4, 6, 8, 10}
⑦Search the best learning rate by grid search in {5e-4, 1e-5, 5e-6, 1e-6, 5e-7, 1e-7}
⑧Cross vadilation: 5 fold
⑨Epoch: 15 for ABCD and UKB, 30 for HCP
2.4.2. Comparative Experiment
①Comparison table:
(他这个不是精度我现在不能用脑子检索出别的文章的AUR导致我不太能现在给出比较结论)
2.4.3. Ablation Study
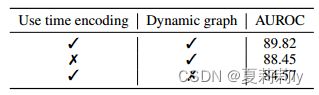
①They remove GRU and change dynamic embedding to static respectively:
②Test of hidden size and layers:
2.5. Conclusion
Their TENET provide valuable interpretability
3. Reference List
Kim B. et al. (2023) 'Large-scale Graph Representation Learning of Dynamic Brain Connectome with Transformers', NeurIPS. doi: https://doi.org/10.48550/arXiv.2312.14939
![[论文精读]Large-scale Graph Representation Learning of Dynamic Brain Connectome with Transformers_第1张图片](http://img.e-com-net.com/image/info8/4a5fe2a3957d48b881b5a442fc185a56.jpg)
![[论文精读]Large-scale Graph Representation Learning of Dynamic Brain Connectome with Transformers_第2张图片](http://img.e-com-net.com/image/info8/0021736ef4f649bcb6a2a8be21b41ab8.jpg)
![[论文精读]Large-scale Graph Representation Learning of Dynamic Brain Connectome with Transformers_第3张图片](http://img.e-com-net.com/image/info8/ecddffa73fd8483487d416295f2f2ac6.jpg)
![[论文精读]Large-scale Graph Representation Learning of Dynamic Brain Connectome with Transformers_第4张图片](http://img.e-com-net.com/image/info8/2ffe4338533241eea1301ac75a7bfbc3.jpg)
![[论文精读]Large-scale Graph Representation Learning of Dynamic Brain Connectome with Transformers_第5张图片](http://img.e-com-net.com/image/info8/85aa43f3f4444638b5c02842ab0552b7.jpg)
![[论文精读]Large-scale Graph Representation Learning of Dynamic Brain Connectome with Transformers_第6张图片](http://img.e-com-net.com/image/info8/a9e5d7122c74493b90186f313b5784d2.jpg)