HDU-1560 DNA sequence(IDA*)
DNA sequence
Time Limit: 15000/5000 MS (Java/Others) Memory Limit: 32768/32768 K (Java/Others)
Problem Description
The twenty-first century is a biology-technology developing century. We know that a gene is made of DNA. The nucleotide bases from which DNA is built are A(adenine), C(cytosine), G(guanine), and T(thymine). Finding the longest common subsequence between DNA/Protein sequences is one of the basic problems in modern computational molecular biology. But this problem is a little different. Given several DNA sequences, you are asked to make a shortest sequence from them so that each of the given sequence is the subsequence of it.
For example, given "ACGT","ATGC","CGTT" and "CAGT", you can make a sequence in the following way. It is the shortest but may be not the only one.
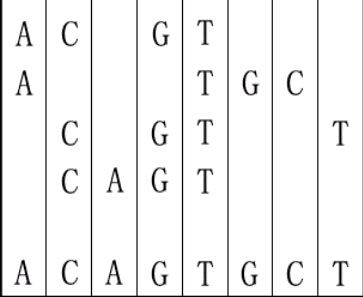
For example, given "ACGT","ATGC","CGTT" and "CAGT", you can make a sequence in the following way. It is the shortest but may be not the only one.

Input
The first line is the test case number t. Then t test cases follow. In each case, the first line is an integer n ( 1<=n<=8 ) represents number of the DNA sequences. The following k lines contain the k sequences, one per line. Assuming that the length of any sequence is between 1 and 5.
Output
For each test case, print a line containing the length of the shortest sequence that can be made from these sequences.
Sample Input
1 4 ACGT ATGC CGTT CAGT
Sample Output
8
数据不是很大,AC后又进入前25了。
开始TLE 5次,一直以为是自己的估价函数没写好,不停的改成现在这个样子,结果还是TLE,百度到大神的代码后发现他的优化比我还少,就想到了一定是初始化的问题,改过之后立刻AC,debug真是一个艰辛的过程。
经过测试后,发现如果只有以DNA最大剩余长度为估价函数返回值,几乎要TLE,所以主要起作用的是剩余DNA中ACGT分别出现的次数的最大值的和
#include <cstdio>
#include <cstring>
#include <algorithm>
using namespace std;
char s[41],dna[9][6],nuc[]={"ACGT"};
int n,len[9],num[9],pre[41][9],step,depth,tp;
int vis[255],mx[255],mxl;
inline bool judge() {//判断是否全部完成匹配
for(int i=0;i<n;++i)
if(len[i]!=num[i])
return false;
return true;
}
int get_h() {//估价函数:返回(未匹配DNA的最大长度)与(剩余DNA中ACGT分别出现的次数的最大值的和)的最大值
int i,j;
mx['A']=mx['C']=mx['G']=mx['T']=mxl=0;
for(i=0;i<n;++i) {
vis['A']=vis['C']=vis['G']=vis['T']=0;
for(j=num[i];j<len[i];++j)
++vis[dna[i][j]];//每一个剩余DNA中ACGT分别出现的次数
mx['A']=max(mx['A'],vis['A']);
mx['C']=max(mx['C'],vis['C']);
mx['G']=max(mx['G'],vis['G']);//剩余DNA中ACGT分别出现的次数的最大值
mx['T']=max(mx['T'],vis['T']);
mxl=max(mxl,len[i]-num[i]);//DNA剩余的长度的最大值
}
return max(mx['A']+mx['C']+mx['G']+mx['T'],mxl);
}
bool IDAstar() {
if(judge())
return true;
if(step+get_h()>=depth)//剪枝
return false;
int i,j;
for(i=0;i<4;++i) {
s[step]=nuc[i];
for(j=0;j<n;++j) {
if(s[step]==dna[j][num[j]])
num[j]+=(pre[step][j]=1);//DNA成功匹配一个字符
else
pre[step][j]=0;
tp+=pre[step][j];//DNA成功匹配字符的个数
}
if(tp!=0) {//DNA至少有一个字符成功匹配时才进入下一层
++step;
if(IDAstar())
return true;
--step;
}
for(j=0;j<n;++j)//DNA成功匹配的字符数复原之字符串到位置step的状态
num[j]-=pre[step][j];
}
return false;
}
int main() {
int i,T;
scanf("%d",&T);
while(T--) {
scanf("%d",&n);
depth=1;
for(i=0;i<n;++i) {
scanf("%s",dna[i]);
depth=max(depth,len[i]=strlen(dna[i]));
num[i]=0;
}
step=0;
while(!IDAstar()) {
++depth;
}
printf("%d\n",step);
}
return 0;
}