此类文章全是笔记,用于帮助自己快速回忆起书籍所讲述的内容,对他人参考意义较少
通识
数据收集、数据存储、数据建模、数据分析、数据变现
数据是承载一定信息的符号
信息是被消除的不确定性
泊松分布
伯努利分布
信息量的计算 I = log2m、H(X)=-log2P
香农公式
回归
回归这种模型只能用来预测和自变量统计的区间比较近的自变量对应的函数值
回归也应该算是分类算法,不同的是,在回归中研究的都是具体的数值,二分类算法不一定,也可以是枚举值或者文本等
线性拟合
import numpy as np
import matplotlib.pyplot as plt
x = [1, 2, 3, 4, 5, 6, 7, 8, 9]
y = [0.199, 0.389, 0.580, 0.783, 0.980, 1.177, 1.380, 1.575, 1.771]
A = np.vstack([x, np.ones(len(x))]).T
a, b = np.linalg.lstsq(A, y, rcond = None)[0] #最小二乘法
x = np.array(x)
y = np.array(y)
plt.plot(x, y, 'o', label='Original data', markersize=10)
plt.plot(x, a*x+b, 'r', label='Fitted line')
plt.legend()
plt.show()
曲线拟合转化为线性拟合
import numpy as np
from scipy.optimize import curve_fit
import matplotlib.pyplot as plt
T = [1960, 1961, 1962, 1963, 1964, 1965, 1966, 1967, 1968]
S = [29.72, 30.61, 31.51, 32.13, 32.34, 32.85, 33.56, 34.20, 34.83]
xdata = np.array(T)
ydata = np.log(np.array(S))
def func(x, a, b):
return a + b*x
popt, pcov = curve_fit(func, xdata, ydata) #非线性最小二乘法拟合函数
plt.plot(xdata, ydata, 'ko', label='Original Data')
plt.plot(xdata, func(xdata, *popt), 'r', label='Fitted Curve')
plt.legend()
plt.show()
聚类 - 连续变量预测
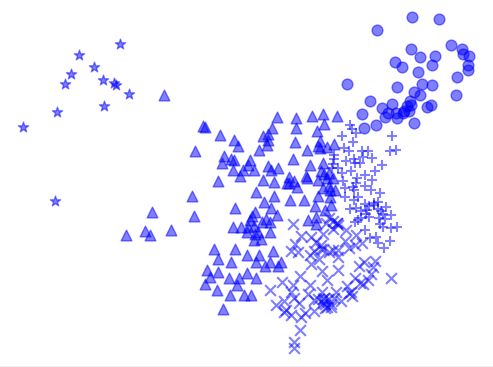
K-Means算法 city.txt
欧式距离度量需要先进行标准化或归一化,同时距离差异越大,个体间差异越大
空间向量余弦夹角的相识度度量不会受指标刻度的影响,余弦值落于区间[-1,1]上,值越大,差异越小
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cluster import KMeans
X = []
with open('./city.txt', 'r', errors='ignore') as f:
for line in f.readlines():
X.append([float(line.split(' ')[2]), float(line.split(' ')[3])])
X = np.array(X)
n_clusters = 5
cls = KMeans(n_clusters).fit(X)
markers = ['^','x','o','*','+']
for i in range(n_clusters):
members = cls.labels_ == i
plt.scatter(X[members,0], X[members,1], s=60, marker = markers[i], c= 'b', alpha =0.5)
plt.title('')
plt.show()
层次聚类
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cluster import AgglomerativeClustering
X = []
with open('./city.txt', 'r', errors='ignore') as f:
for line in f.readlines():
X.append([float(line.split(' ')[2]), float(line.split(' ')[3])])
X = np.array(X)
n_clusters = 5
cls = AgglomerativeClustering(linkage='ward',n_clusters=n_clusters).fit(X)
markers = ['^','x','o','*','+']
for i in range(n_clusters):
members = cls.labels_ == i
plt.scatter(X[members,0], X[members,1], s=60, marker = markers[i], c= 'b', alpha =0.5)
plt.title('')
plt.show()
密度聚类
带拐弯的、狭长、不规则形状的聚类效果比K-均值聚类要好
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cluster import DBSCAN
X = [
[9670250, 1392358258], #中国
[2960000, 1247923065], #印度
[9629091, 317408015], #美国
[8514877, 201032714], #巴西
[377873, 127280000], #日本
[7692024, 23540517], #澳大利亚
[17075400, 143551289], #俄罗斯
[513115, 67041000], #泰国
[181035, 14805358], #柬埔寨
[99600, 50400000], #韩国
[120538, 24052231] #朝鲜
]
X = np.array(X)
# 归一化 将不同维度的数据都投影到以10 000为最大值的正方形区域里,防止因为不同维度的距离比重过大,失去聚类的意义
a = X[:,:1] / 17075400.0 * 10000
b = X[:,1:] / 1392358258.0 * 10000
X = np.concatenate((a,b),axis=1)
cls = DBSCAN(eps=2000,min_samples=1).fit(X)
n_clusters = len(set(cls.labels_))
markers = ['^','x','o','*','+']
for i in range(n_clusters):
members = cls.labels_ == i
plt.scatter(X[members,0], X[members,1], s=60, marker = markers[i], c= 'b', alpha =0.5)
plt.title('DBSCAN')
plt.show()
聚类评估
1.聚类趋势-必须是非随机样本
用霍普金斯统计量来进行量化评估
如果整个样本空间没有聚类趋势或者不明显,那么H应该位0.5左右,否则应该接近与1
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cluster import DBSCAN
X = [
[9670250, 1392358258], #中国
[2960000, 1247923065], #印度
[9629091, 317408015], #美国
[8514877, 201032714], #巴西
[377873, 127280000], #日本
[7692024, 23540517], #澳大利亚
[17075400, 143551289], #俄罗斯
[513115, 67041000], #泰国
[181035, 14805358], #柬埔寨
[99600, 50400000], #韩国
[120538, 24052231] #朝鲜
]
X = np.array(X)
# 归一化
a = X[:,:1] / 17075400.0 * 10000
b = X[:,1:] / 1392358258.0 * 10000
X = np.concatenate((a,b),axis=1)
pn = X[np.random.choice(X.shape[0],3,replace = False),:]
xn = []
for i in pn:
distance_min = 1000000
for j in X:
if np.array_equal(j,i):
continue
distance = np.linalg.norm(j-1)
if distance_min > distance:
distance_min = distance
xn.append(distance_min)
qn = X[np.random.choice(X.shape[0],3,replace = False),:]
yn = []
for i in qn:
distance_min = 1000000
for j in X:
if np.array_equal(j,i):
continue
distance = np.linalg.norm(j-1)
if distance_min > distance:
distance_min = distance
yn.append(distance_min)
H = float(np.sum(yn)) / (np.sum(xn) + np.sum(yn))
print(H) #在0.5附近
2.簇数确定-肘方法
3.聚类质量-轮廓系数
import numpy as np
import matplotlib.pyplot as plt
from sklearn.cluster import KMeans
X = [
[9670250, 1392358258], #中国
[2960000, 1247923065], #印度
[9629091, 317408015], #美国
[8514877, 201032714], #巴西
[377873, 127280000], #日本
[7692024, 23540517], #澳大利亚
[17075400, 143551289], #俄罗斯
[513115, 67041000], #泰国
[181035, 14805358], #柬埔寨
[99600, 50400000], #韩国
[120538, 24052231] #朝鲜
]
X = np.array(X)
# 归一化
a = X[:,:1] / 17075400.0 * 10000
b = X[:,1:] / 1392358258.0 * 10000
X = np.concatenate((a,b),axis=1)
n_clusters = 3
cls = KMeans(n_clusters).fit(X)
def manhattan_distance(x,y):
return np.sum(abs(x-y))
distance_sum = 0
for v in X[1:]:
distance_sum += manhattan_distance(np.array(X[0]),np.array(v))
av = distance_sum / len(X[1:])
print(av)
distance_min = 100000
for i in range(n_clusters):
group = cls.labels_ == i
members = X[group, :]
for v in members:
if np.array_equal(v,X[0]):
continue
distance = manhattan_distance(np.array(v),cls.cluster_centers_)
if distance_min > distance:
distance_min = distance
bv = distance_sum / n_clusters
print(bv)
sv = float(bv-av)/max(av,bv)
print(sv) #0.7
分类 - 离散变量预测
朴素贝叶斯
基本思想
1.已知类条件概率密度参数表达式和先验概率
2.利用贝叶斯公式转换成后验概率
3.根据后验概率大小进行决策分类
from sklearn.naive_bayes import GaussianNB
#0:晴 1:阴 2:降水 3:多云
data_table = [
['date', 'weather'],
[1, 0],
[2, 1],
[3, 2],
[4, 1],
[5, 2],
[6, 0],
[7, 0],
[8, 3],
[9, 1],
[10, 1]
]
X = [[0],[1],[2],[1],[2],[0],[0],[3],[1]]
Y = [1,2,1,2,0,0,3,1,1]
cls = GaussianNB().fit(X,Y)
p = [[1]]
print(cls.predict(p)) #[[2]]
from sklearn.naive_bayes import GaussianNB
# 基因片段A 基因片段B 高血压 胆结石
# 0:否 1:是
data_table = [
[1, 1, 1, 0],
[0, 0, 0, 1],
[0, 1, 0, 0],
[1, 0, 0, 0],
[1, 1, 0, 1],
[1, 0, 0, 1],
[0, 1, 1, 1],
[0, 0, 0, 0],
[1, 0, 1, 0],
[0, 1, 0, 1]
]
# 基因片段
X = [[1, 1], [0, 0], [0, 1], [1, 0], [1, 1], [1, 0], [0, 1], [0, 0], [1, 0], [0, 1]]
# 高血压
Y1 = [1, 0, 0, 0, 0, 0, 1, 0, 1, 0]
cls = GaussianNB().fit(X,Y1)
p1 = [[1,1]]
print(cls.predict(p1)) #[1]
# 胆结石
Y2 = [0, 1, 0, 0, 1, 1, 1, 0, 0, 1]
cls = GaussianNB().fit(X,Y2)
p2 = [[1,1]]
print(cls.predict(p1)) #[0]
决策树归纳
计算熵
import math
# 大专 本科 硕士
education = (2.0/12, 5.0/12, 5.0/12)
# 大专
junior_college=(1.0/2, 1.0/2)
# 本科
undergraduate = (3.0/5, 1.0/5)
# 硕士
master = (4.0/5, 1.0/5)
date_per = (junior_college, undergraduate, master)
# 相亲字段下划分的熵
def info_date(p):
info = 0
for v in p:
info += v*math.log(v,2)
return info
# 学历字段下划分的熵
def infoA():
info = 0
for i in range(len(education)):
info += -education[i] * info_date(date_per[i])
return info
print(infoA()) #0.8452054459852327
# 年龄
age = [25,25,28,28,29,30,33,34,35,36,40,46]
date = [0,1,1,0,1,1,1,1,1,0,0,0]
# 需要划分n-1次 这里取28 29
# 左右占总比百分比
split_per = (4.0/12, 8.0/12)
# 左分类百分比
date_left = (1.0/2, 1/2)
# 右分类百分比
date_right = (5.0/8, 3.0/8)
date_per = (date_left,date_right)
# 相亲字段下划分的熵
def info_date(p):
info = 0
for v in p:
info += v*math.log(v,2)
return info
def infoA():
info = 0
for i in range(len(split_per)):
info += -split_per[i] * info_date(date_per[i])
return info
print(infoA()) #0.9696226686166431
构造
1.找到信息增益最大的字段A和切分点v
2.决定根节点的字段A和切分点v
3.把字段A拿走,再从第一步开始找
随机深林
from sklearn.ensemble import RandomForestClassifier
# 学历 0 大专 1本科 2硕士
# 年龄 身高 年收入 学历
X = [
[25, 179, 15, 0],
[33, 190, 19, 0],
[28, 180, 18, 2],
[25, 178, 18, 2],
[46, 100, 100, 2],
[40, 170, 170, 1],
[34, 174, 20, 2],
[35, 170, 25, 2],
[30, 180, 35, 1],
[28, 174, 30, 1],
[29, 176, 36, 1],
[36, 181, 55, 1]
]
#相亲 0否 1是
Y = [0, 1, 1, 1, 0, 0, 1, 0, 1, 1, 0, 1]
clf = RandomForestClassifier().fit(X, Y)
p = [[28 ,180 ,18 ,2]]
print(clf.predict(p)) #1
隐马尔科夫模型
1.维特比算法
知道隐含状态数量、转换概率、可见状态链求隐含状态链
import numpy as np
import math
jin = ['近', '斤', '今', '金', '尽']
jin_per = [0.3, 0.2, 0.1, 0.06, 0.03]
jintian = ['天', '填', '田', '甜', '添']
jintian_per = [
[0.001, 0.001, 0.001, 0.001, 0.001],
[0.001, 0.001, 0.001, 0.001, 0.001],
[0.990, 0.001, 0.001, 0.001, 0.001],
[0.002, 0.001, 0.850, 0.001, 0.001],
[0.001, 0.001, 0.001, 0.001, 0.001]
]
wo = ['我', '窝', '喔', '握', '卧']
wo_per = [0.400, 0.150, 0.090, 0.050, 0.030]
women = ['们', '门', '闷', '焖', '扪']
women_per = [
[0.970, 0.001, 0.003, 0.001, 0.001],
[0.001, 0.001, 0.001, 0.001, 0.001],
[0.001, 0.001, 0.001, 0.001, 0.001],
[0.001, 0.001, 0.001, 0.001, 0.001],
[0.001, 0.001, 0.001, 0.001, 0.001]
]
N = 5
def found_from_oneword(oneword_per):
index = []
values = []
a = np.array(oneword_per)
for v in np.argsort(a)[::-1][:N]:
index.append(v)
values.append(oneword_per[v])
return index, values
def found_from_twoword(oneword_per, twoword_per):
last = 0
for i in range(len(jin_per)):
current = np.multiply(oneword_per[i], twoword_per[i])
if i == 0:
last = current
else:
last = np.concatenate((last, current), axis = 0)
index = []
values = []
for v in np.argsort(last)[::-1][:N]:
index.append([math.floor(v / 5), v % 5])
values.append(last[v])
return index, values
def predict(word):
if word == 'jin':
for i in found_from_oneword(jin_per)[0]:
print(jin[i])
elif word == 'jintian':
for i in found_from_twoword(jin_per, jintian_per)[0]:
print(i)
print(jin[i[0]] + jintian[i[1]])
elif word == 'wo':
for i in found_from_oneword(wo_per)[0]:
print(wo[i])
elif word == 'women':
for i in found_from_twoword(wo_per, women_per)[0]:
print(wo[i[0]] + women[i[1]])
elif word == 'jintianwo':
index1, values1 = found_from_oneword(wo_per)
index2, values2 = found_from_twoword(jin_per, jintian_per)
last = np.multiply(values1, values1)
for v in np.argsort(last)[::-1][:N]:
print(jin[index2[v][0]], jintian[index2[v][1]], wo[v])
elif word == 'jintianwomen':
index1, values1 = found_from_twoword(jin_per, jintian_per)
index2, values2 = found_from_twoword(wo_per,women_per)
last = np.multiply(values1, values1)
for v in np.argsort(last)[::-1][:N]:
print(jin[index1[v][0]], jintian[index1[v][1]], wo[index2[v][0]], women[index2[v][0]])
if __name__ == '__main__':
predict('jin') #近,斤,今,金,尽
predict('jintian') #今天,金田 ......
predict('jintianwo') #今天我 ......
predict('jintianwomen') #今天我们 ......
2.前向算法
知道隐含状态数量,转换概率、可见状态链求结果概率
比较两个序列概率的大小
支持向量机
from sklearn import svm
# 年龄
X = [[34],[33],[32],[31],[30],[30],[25],[23],[22],[18]]
# 质量
Y = [1,0,1,0,1,1,0,1,0,1]
clf = svm.SVC(kernel='rbf').fit(X,Y)
P = [[26]]
print(clf.predict(P)) #[1]
遗传算法
import random
import math
import numpy as np
# 染色体长度
CHROMOSOME_SIZE = 15
# 收敛条件、判断退出
def is_finished(last_three):
s = sorted(last_three)
if s[0] and s[2] - s[0] < 0.01 * s[0]:
return True
else:
return False
# 初始化染色体样态
def init():
chromosome_state1 = ['000000100101001', '101010101010101']
chromosome_state2 = ['011000100101100', '001100110011001']
chromosome_state3 = ['001000100100101', '101010101010101']
chromosome_state4 = ['000110100100100', '110011001100110']
chromosome_state5 = ['100000100100101', '101010101010101']
chromosome_state6 = ['101000100100100', '111100001111000']
chromosome_state7 = ['101010100110100', '101010101010101']
chromosome_state8 = ['100110101101000', '000011110000111']
chromosome_states = [
chromosome_state1,
chromosome_state2,
chromosome_state3,
chromosome_state4,
chromosome_state5,
chromosome_state6,
chromosome_state7,
chromosome_state8
]
return chromosome_states
# 计算适应度
def fitness(chromosome_states):
fitnesses = []
for chromosome_state in chromosome_states:
if chromosome_state[0][0] == '1':
x = 10 * (-float(int(chromosome_state[0][1:], 2) - 1) / 16384)
else:
x = 10 * (-float(int(chromosome_state[0], 2) + 1) / 16384)
if chromosome_state[1][0] == '1':
y = 10 * (-float(int(chromosome_state[1][1:], 2) - 1) / 16384)
else:
y = 10 * (-float(int(chromosome_state[1], 2) + 1) / 16384)
z = y * math.sin(x) + x * math.cos(y)
fitnesses.append(z)
return fitnesses
# 筛选
def filter(chromosome_states, fitnesses):
# 重量大于80的被淘汰
chromosome_states_new = []
top1_fitness_index = 0
for i in np.argsort(fitnesses)[::-1][:8].tolist():
chromosome_states_new.append(chromosome_states[i])
top1_fitness_index = i
return chromosome_states_new, top1_fitness_index
# 产生下一代
def crossover(chromosome_states):
chromosome_states_new = []
index = len(chromosome_states) - 1
while chromosome_states:
chromosome_state = chromosome_states.pop(0)
for v in chromosome_states:
pos = random.choice(range(8, CHROMOSOME_SIZE - 1))
chromosome_states_new.append([chromosome_state[0][:pos] + v[0][pos:], chromosome_state[1][:pos] + v[1][pos:]])
chromosome_states_new.append([chromosome_state[1][pos:] + v[0][:pos], chromosome_state[1][pos:] + v[0][:pos]])
return chromosome_states_new
# 基因突变
def mutation(chromosome_states):
n = int(5.0 / 100 * len(chromosome_states))
while n > 0:
n -= 1
chromosome_state = random.choice(chromosome_states)
index = chromosome_states.index(chromosome_state)
pos = random.choice(range(len(chromosome_state)))
x = chromosome_state[0][:pos] + str(int(not int(chromosome_state[0][pos]))) + chromosome_state[0][pos+1:]
y = chromosome_state[1][:pos] + str(int(not int(chromosome_state[1][pos]))) + chromosome_state[1][pos+1:]
chromosome_states[index] = [x, y]
if __name__ == '__main__':
chromosome_states = init()
last_three = [0] * 3
last_num = 0
n = 100
while n > 0:
n -= 1
chromosome_states = crossover(chromosome_states)
mutation(chromosome_states)
fitnesses = fitness(chromosome_states)
chromosome_states, top1_fitness_index = filter(chromosome_states, fitnesses)
last_three[last_num] = fitnesses[top1_fitness_index]
if is_finished(last_three):
break
if last_num >= 2:
last_num = 0
else:
last_num += 1
关联分析
支持度和置信度
Apriori算法
用户画像
推荐算法
贝叶斯分类
搜索记录
User-based CF 基于用户的协同过滤
Item-based CF 基于商品的协同过滤
文本挖掘
Rocchio算法
缺陷
1. 一个类别的文档仅仅在一个质心周围
2.训练数据绝对正确
朴素贝叶斯算法
from sklearn.datasets import fetch_20newsgroups
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfTransformer
from sklearn.naive_bayes import MultinomialNB
from pprint import pprint
# newsgroups_train = fetch_20newsgroups(subset = 'train')
# pprint(list(newsgroups_train.target_names))
categories = ['alt.atheism', 'comp.graphics', 'sci.med', 'soc.religion.christian']
twenty_train = fetch_20newsgroups(subset = 'train', categories = categories)
count_vect = CountVectorizer()
tfidf_transformer = TfidfTransformer()
X_train_counts = count_vect.fit_transform(twenty_train.data)
X_train_tfidf = tfidf_transformer.fit_transform(X_train_counts)
clf = MultinomialNB().fit(X_train_tfidf, twenty_train.target)
docs_new = ['God is love', 'OpenGL on the GPU is fast']
X_new_counts = count_vect.transform(docs_new)
X_new_tfidf = tfidf_transformer.transform(X_new_counts)
predicted = clf.predict(X_new_tfidf)
for doc, category in zip(docs_new, predicted):
print('%r => %s' % (doc, twenty_train.target_names[category]) )
# 'God is love' => soc.religion.christian
# 'OpenGL on the GPU is fast' => comp.graphics
K-近邻算法
优点 - 可以克服Rocchio算法无法处理线性的缺陷,训练成本非常低
缺点 - 计算成本高
from sklearn.datasets import fetch_20newsgroups
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfTransformer
from sklearn.neighbors import KNeighborsClassifier
categories = ['alt.atheism', 'comp.graphics', 'sci.med', 'soc.religion.christian']
twenty_train = fetch_20newsgroups(subset = 'train', categories = categories)
count_vect = CountVectorizer()
tfidf_transformer = TfidfTransformer()
X_train_counts = count_vect.fit_transform(twenty_train.data)
X_train_tfidf = tfidf_transformer.fit_transform(X_train_counts)
clf = KNeighborsClassifier().fit(X_train_tfidf, twenty_train.target)
docs_new = ['God is love', 'OpenGL on the GPU is fast']
X_new_counts = count_vect.transform(docs_new)
X_new_tfidf = tfidf_transformer.transform(X_new_counts)
predicted = clf.predict(X_new_tfidf)
for doc, category in zip(docs_new, predicted):
print('%r => %s' % (doc, twenty_train.target_names[category]) )
# 'God is love' => soc.religion.christian
# 'OpenGL on the GPU is fast' => comp.graphics
支持向量机SVM算法 - SVM分类器的文本分类效果很好
优点 - 通用性较好、分类精度高、分类速度快、分类速度与训练样本个数无关、精度和召回率优于KNN及朴素贝叶斯
缺点 - 训练速度受训练集规模影响、计算开销较大
from sklearn.datasets import fetch_20newsgroups
from sklearn.feature_extraction.text import CountVectorizer
from sklearn.feature_extraction.text import TfidfTransformer
from sklearn import svm
categories = ['alt.atheism', 'comp.graphics', 'sci.med', 'soc.religion.christian']
twenty_train = fetch_20newsgroups(subset = 'train', categories = categories)
count_vect = CountVectorizer()
tfidf_transformer = TfidfTransformer()
X_train_counts = count_vect.fit_transform(twenty_train.data)
X_train_tfidf = tfidf_transformer.fit_transform(X_train_counts)
clf = svm.SVC(kernel='linear').fit(X_train_tfidf, twenty_train.target)
docs_new = ['God is love', 'OpenGL on the GPU is fast']
X_new_counts = count_vect.transform(docs_new)
X_new_tfidf = tfidf_transformer.transform(X_new_counts)
predicted = clf.predict(X_new_tfidf)
for doc, category in zip(docs_new, predicted):
print('%r => %s' % (doc, twenty_train.target_names[category]) )
# 'God is love' => soc.religion.christian
# 'OpenGL on the GPU is fast' => comp.graphics
人工神经网络
FANN库
BP神经网络
玻尔兹曼机
卷积神经网络