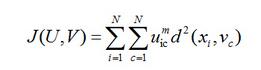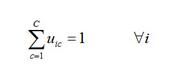FCM 算法
一 FCM算法概述
FCM算法的全称是模糊C均值聚类算法,和K-means算法同属于聚类算法,但却有着本质的区别,就其命名而言,模糊二字无疑是该算法的重点,下面就先简单介绍一下:
隶属度和模糊集
隶属度函数用来描述元素x属于一个集合B的程度,假定为UB(x),其中x为B中的任意元素,UB(x)的取值范围为[0,1]。在隶属度函数的基础上,称空间上X={x}上的隶属度函数为一个模糊集合。
模糊聚类分析
传统的聚类分析是把每个元素严格的划分到一个类中,属于硬划分。模糊聚类分析将聚类生成的每个簇均看做模糊集合,通过隶属度来确定聚类关系,是一种柔性划分,得到元素属于各个簇的不确定性程度,使得聚类结果更加准确灵活,因此,模糊聚类分析逐渐成为聚类分析的主流。
3.FCM
FCM算法是将N个L维向量分为C个模糊组,通过迭代不断更新隶属度以及聚类中心,最小化目标函数对数据进行聚类。
这里,m指的是模糊加权系数,它的值大于1;d(xi,vc)表示的是第i个数据点与第c个聚类中心的欧式距离;uic是隶属度矩阵中的元素,且[0,1];vc是对应于每个聚类的聚类中心。
为了求含有约束条件的目标函数的极值,引入拉格朗日因子构造新的目标函数:

对于目标函数求极值的最优化条件如下:

根据上述公式不断迭代求出满足条件的隶属度以及聚类中心。具体的FCM算法步骤如下:
首先,给定一个由N个L维向量组成的数据集X以及所要分得的类别个数C,自定义隶属度矩阵
(1)设定类别的个数C和模糊系数m;
(2)‚初始化隶属度矩阵且满足公式(2)中的归一化条件;
(3)根据公式(5)计算聚类中心;
(4)根据公式(4)更新隶属度矩阵;
(5)根据矩阵范数比较迭代的隶属度矩阵,如果![]() ,迭代停止,否则返回(3)。
,迭代停止,否则返回(3)。
二 MATLAB实现
根据上述公式及步骤,采用MATLAB实现,具体如下:
(1)选取UCI数据库中的wine数据实现,代码:
clear all
clc;
%% 导入数据
load wine.txt;
cluster_n=6;
data = wine;
data(:,1) = [];
data(:,size(data,2)) = [];
data_n = size(data, 1); %数据的个数
in_n = size(data, 2);% 数据维数
%% 定义变量
default_options = [10; % 隶属度函数的幂次方
300; % 最大迭代次数
1e-5; %步长
1]; % 判定条件
options = default_options;
expo = options(1); % Exponent for U 隶属度函数的幂次方
max_iter = options(2); % Max. iteration 最大迭代次数
min_impro = options(3); % Min. improvement 最小进化步长
display = options(4); % Display info or not 显示信息与否
obj_fcn = zeros(max_iter, 1); % Array for objective function
tic
%% 初始化隶属度矩阵并归一
U = rand(cluster_n, data_n); %rand()产生随机矩阵
col_sum = sum(U);
U = U./col_sum(ones(cluster_n, 1), :);%归一化
%% 开始迭代
for i = 1:max_iter,%迭代次数控制
tic
mf = U.^expo; % MF matrix after exponential modification
center = mf*data./((ones(size(data, 2), 1)*sum((mf')))'); % 建立新的聚类中心
out = zeros(size(center, 1), size(data, 1)); %每个点到每个中心的距离,行数为中心数
if size(center, 2) > 1,%样本的维数大于一执行以下程序
for k = 1:size(center, 1),%给K赋值
abc = ((data-ones(size(data,1),1) * center(k,:)).^2)';
out(k, :) = sqrt(sum(abc));%得到欧氏距离
end
else % 1-D data
for k = 1:size(center, 1),
out(k, :) = abs(center(k)-data)';
end
end
obj_fcn(i) = sum(sum((out.^2).*mf)); % 目标函数
tmp = out.^(-2/(expo-1)); % 根据新的隶属度矩阵求解公式求出
U= tmp./(ones(cluster_n, 1)*sum(tmp)); % 新的隶属度矩阵
if display,
fprintf('Iteration count = %d, obj. fcn = %f\n', i, obj_fcn(i));
%输出迭代次数和函数的结果
end
% check termination condition
if i > 1, %进化步长控制
if abs(obj_fcn(i) - obj_fcn(i-1)) < min_impro, break; end,
end
toc
end
toc
plot(U(1,:),'-ro');
grid on
hold on
plot(U(2,:),'-g*');
plot(U(3,:),'-b+');
ylabel('Membership degrees')
legend('FCM1','FCM2','FCM3','location','northeast');
toc结果如下:
(2)选取一张MRI图像,进行图像分割,代码如下:
clear all
clc;
a=imread('MRI.jpg');
I=imnoise(a,'salt & pepper',0.05);
figure(1);
imshow(I);title('加噪图像');
[height,width,c]=size(a);
if c~=1
a=rgb2gray(a);
end
a=double(a);
[row,column]=size(a);
data = a(:);
data_n = size(data,1);
cluster_num = 4;
default_options = [2.0; % 隶属度函数的幂次方
300; % 最大迭代次数
1e-5; %步长
1]; % 判定条件
options = default_options;
expo = options(1); % Exponent for U 隶属度函数的幂次方
max_iter = options(2); % Max. iteration 最大迭代次数
min_impro = options(3); % Min. improvement 最小进化步长
display = options(4); % Display info or not 显示信息与否
obj_fcn = zeros(max_iter, 1); % Array for objective function
membership = zeros(height,width,cluster_num);
center = zeros(cluster_num,1);
tic
% 初始化隶属度并归一
for i=1:height
for j=1:width
member_sum=0;
for k=1:cluster_num
membership(i,j,k)=rand();
member_sum = member_sum + membership(i,j,k);
end
for p =1:cluster_num
membership(i,j,p) = membership(i,j,p) / member_sum;
end
end
end
tic
for i = 1:max_iter,%迭代次数控制
mf = membership.^expo;
%%%%%%%%建立聚类中心
for m = 1:cluster_num
to = 0;
tp =0;
for j = 1:height
for t = 1:width
to = to + membership(j,t,m) * a(j,t);
tp = tp + membership(j,t,m);
end
end
center(m,1) = to / tp;
end
%%%%%%%得到欧式距离以及目标函数
out = zeros(height,width,cluster_num);
for m =1:height
for j =1:width
for t = 1:cluster_num
out(m,j,t) = abs(a(m,j) - center(t,1));
obj_fcn(i) = obj_fcn(i) + (membership(m,j,t).^expo) * (out(m,j,t).^2);
end
end
end
for m = 1:height
for j = 1:width
for r = 1:cluster_num
top =0;
for t = 1:cluster_num
top = top + (out(m,j,r) / out(m,j,t)).^(expo - 1);
end
membership(m,j,r) = 1 / top;
end
end
end
%%%%%%归一化隶属度
for m=1:height
for j = 1:width
member_sum = 0;
for k = 1:cluster_num
member_sum = member_sum + membership(m,j,k);
end
for p = 1:cluster_num
membership(m,j,p) = membership(m,j,p) / member_sum;
end
end
end
if display,
fprintf('Iteration count = %d, obj. fcn = %f\n', i, obj_fcn(i));
%输出迭代次数和函数的结果
end
% check termination condition
if i > 1, %进化步长控制
if abs(obj_fcn(i) - obj_fcn(i-1)) < min_impro, break; end,
end
end
toc
%%%%%%证得如自定义图像中的MCR不能计算,故在此继续尝试直接用newing和A相比较
A = ones(height,width,1);
for i = 1:height
for j = 1:width
if (fix(a(i,j) / 85) == 1)
A(i,j,1) = 2;
end
if (fix(a(i,j) / 85) == 2)
A(i,j,1) = 3;
end
if (fix(a(i,j,1) / 85) == 3)
A(i,j,1) = 4;
end
end
end
A = reshape(A,1,data_n);
newing = zeros(row,column);
for i=1:row
for j=1:column
maxmembership=membership(i,j,1);
index=1;
for k=2:cluster_num
if(membership(i,j,k)>maxmembership)
maxmembership=membership(i,j,k);
index=k;
end
end
newing(i,j) = round(255 * (1-(index-1)/(cluster_num-1)));
end
end
B = reshape(newing,1,data_n);
b = fix((max(B) - B(1,1)) / cluster_num);
for i = 2:data_n
if B(1,i) == B(1,1)
B(1,i) = 1;
elseif (fix(B(1,i) / b) == 2)
B(1,i) = 2;
elseif (fix(B(1,i) / b) == 3)
B(1,i) = 3;
else
B(1,i) = 4;
end
end
B(1,1) = 1;
sum = 0;
for i = 1:data_n
if ( A(1,i) ~= B(1,i))
sum = sum + 1;
end
end
MCR = sum / data_n;
fprintf('MCR = %d\n',MCR);
S = 0;
for i = 1:data_n
S = S + (A(1,i) - B(1,i)).^2;
end
RMS = sqrt(S / (data_n * (data_n -1)));
fprintf('RMS = %d\n',RMS);
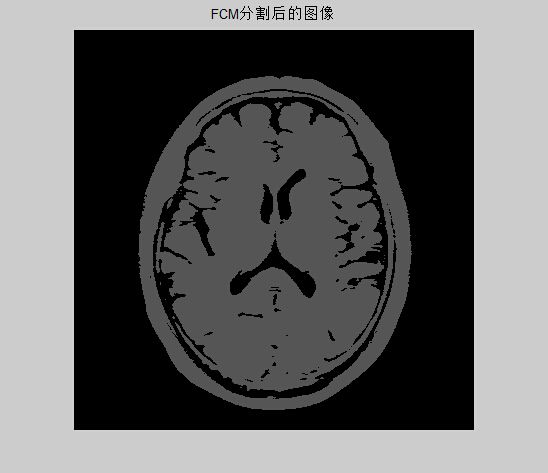
figure(2);
imshow((uint8(newing)));
title('FCM分割后的图像'); 转载自: http://www.cnblogs.com/ybjourney/p/4735335.html