一.相关性分析
1.简介
计算两个系列数据之间的相关性是统计中的常见操作。在spark.ml中提供了很多算法用来计算两两的相关性。目前支持的相关性算法是Pearson和Spearman。Correlation使用指定的方法计算输入数据集的相关矩阵。输出是一个DataFrame,其中包含向量列的相关矩阵。
2.代码实现
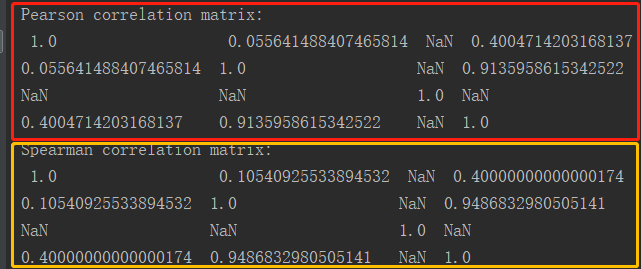
1 package ml 2 3 import org.apache.log4j.{Level, Logger} 4 import org.apache.spark.ml.linalg.{Matrix, Vectors} 5 import org.apache.spark.ml.stat.Correlation 6 import org.apache.spark.sql.{Row, SparkSession} 7 /** 8 * Created by Administrator on 2019/11/28. 9 */ 10 object CorrelationDemo { 11 Logger.getLogger("org").setLevel(Level.WARN) 12 def main(args: Array[String]) { 13 val spark = SparkSession.builder().appName(s"${this.getClass.getSimpleName}").master("local[2]").getOrCreate() 14 import spark.implicits._ // 导入,否则无法使用toDF算子 15 16 val data = Seq( 17 Vectors.sparse(4, Seq((0, 1.0), (3, -2.0))), 18 Vectors.dense(4.0, 5.0, 0.0, 3.0), 19 Vectors.dense(6.0, 7.0, 0.0, 8.0), 20 Vectors.sparse(4, Seq((0, 9.0), (3, 1.0))) 21 ) 22 23 val df = data.map(Tuple1.apply).toDF("features") 24 val Row(coeff : Matrix) = Correlation.corr(df, "features").head 25 println(s"Pearson correlation matrix:\n $coeff") 26 27 df.cache() 28 val Row(coeff2 : Matrix) = Correlation.corr(df, "features", "spearman").head 29 println(s"Spearman correlation matrix:\n $coeff2") 30 } 31 }
3.源码分析
package org.apache.spark.ml.stat import scala.collection.JavaConverters._ import org.apache.spark.annotation.{Experimental, Since} import org.apache.spark.ml.linalg.{SQLDataTypes, Vector} import org.apache.spark.mllib.linalg.{Vectors => OldVectors} import org.apache.spark.mllib.stat.{Statistics => OldStatistics} import org.apache.spark.sql.{DataFrame, Dataset, Row} import org.apache.spark.sql.types.{StructField, StructType} /** * API for correlation functions in MLlib, compatible with DataFrames and Datasets. * * The functions in this package generalize the functions in [[org.apache.spark.sql.Dataset#stat]] * to spark.ml's Vector types. */ @Since("2.2.0") @Experimental object Correlation { /** * :: Experimental :: * Compute the correlation matrix for the input Dataset of Vectors using the specified method. * Methods currently supported: `pearson` (default), `spearman`. * * @param dataset A dataset or a dataframe * @param column The name of the column of vectors for which the correlation coefficient needs * to be computed. This must be a column of the dataset, and it must contain * Vector objects. * @param method String specifying the method to use for computing correlation. * Supported: `pearson` (default), `spearman` * @return A dataframe that contains the correlation matrix of the column of vectors. This * dataframe contains a single row and a single column of name * '$METHODNAME($COLUMN)'. * @throws IllegalArgumentException if the column is not a valid column in the dataset, or if * the content of this column is not of type Vector. * * Here is how to access the correlation coefficient: * {{{ * val data: Dataset[Vector] = ... * val Row(coeff: Matrix) = Correlation.corr(data, "value").head * // coeff now contains the Pearson correlation matrix. * }}} * * @note For Spearman, a rank correlation, we need to create an RDD[Double] for each column * and sort it in order to retrieve the ranks and then join the columns back into an RDD[Vector], * which is fairly costly. Cache the input Dataset before calling corr with `method = "spearman"` * to avoid recomputing the common lineage. */ @Since("2.2.0") def corr(dataset: Dataset[_], column: String, method: String): DataFrame = { val rdd = dataset.select(column).rdd.map { case Row(v: Vector) => OldVectors.fromML(v) } val oldM = OldStatistics.corr(rdd, method) val name = s"$method($column)" val schema = StructType(Array(StructField(name, SQLDataTypes.MatrixType, nullable = false))) dataset.sparkSession.createDataFrame(Seq(Row(oldM.asML)).asJava, schema) } /** * Compute the Pearson correlation matrix for the input Dataset of Vectors. */ @Since("2.2.0") def corr(dataset: Dataset[_], column: String): DataFrame = { corr(dataset, column, "pearson") } }
4.执行结果
二.卡方检验
1.简介
ChiSquareTest针对标签上的每个功能进行Pearson独立性检验。对于每个特征,将(特征,标签)对转换为列矩阵,针对该列矩阵计算卡方统计量。所有标签和特征必须是分类数据。
2.代码实现
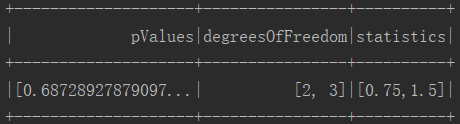
1 package ml 2 3 import org.apache.log4j.{Level, Logger} 4 import org.apache.spark.ml.linalg.Vectors 5 import org.apache.spark.ml.stat.ChiSquareTest 6 import org.apache.spark.sql.SparkSession 7 8 /** 9 * Created by Administrator on 2019/11/28. 10 */ 11 object ChiSquare { 12 Logger.getLogger("org").setLevel(Level.WARN) 13 def main(args: Array[String]) { 14 val spark = SparkSession.builder().appName(s"${this.getClass.getSimpleName}").master("local[2]").getOrCreate() 15 import spark.implicits._// 导入,否则无法使用toDF算子 16 17 val data = Seq( 18 (0.0, Vectors.dense(0.5, 10.0)), 19 (0.0, Vectors.dense(1.5, 20.0)), 20 (1.0, Vectors.dense(1.5, 30.0)), 21 (0.0, Vectors.dense(3.5, 30.0)), 22 (0.0, Vectors.dense(3.5, 40.0)), 23 (1.0, Vectors.dense(3.5, 40.0)) 24 ) 25 26 val df = data.toDF("label", "features") 27 val chi = ChiSquareTest.test(df, "features", "label") // 卡方检验 28 chi.show() 29 } 30 }
3.源码分析
package org.apache.spark.ml.stat import org.apache.spark.annotation.{Experimental, Since} import org.apache.spark.ml.linalg.{Vector, Vectors, VectorUDT} import org.apache.spark.ml.util.SchemaUtils import org.apache.spark.mllib.linalg.{Vectors => OldVectors} import org.apache.spark.mllib.regression.{LabeledPoint => OldLabeledPoint} import org.apache.spark.mllib.stat.{Statistics => OldStatistics} import org.apache.spark.sql.DataFrame import org.apache.spark.sql.functions.col /** * :: Experimental :: * * Chi-square hypothesis testing for categorical data. * * See http://en.wikipedia.org/wiki/Chi-squared_test">Wikipedia for more information * on the Chi-squared test. */ @Experimental @Since("2.2.0") object ChiSquareTest { /** Used to construct output schema of tests */ private case class ChiSquareResult( pValues: Vector, degreesOfFreedom: Array[Int], statistics: Vector) /** * Conduct Pearson's independence test for every feature against the label. For each feature, the * (feature, label) pairs are converted into a contingency matrix for which the Chi-squared * statistic is computed. All label and feature values must be categorical. * * The null hypothesis is that the occurrence of the outcomes is statistically independent. * * @param dataset DataFrame of categorical labels and categorical features. * Real-valued features will be treated as categorical for each distinct value. * @param featuresCol Name of features column in dataset, of type `Vector` (`VectorUDT`) * @param labelCol Name of label column in dataset, of any numerical type * @return DataFrame containing the test result for every feature against the label. * This DataFrame will contain a single Row with the following fields: * - `pValues: Vector` * - `degreesOfFreedom: Array[Int]` * - `statistics: Vector` * Each of these fields has one value per feature. */ @Since("2.2.0") def test(dataset: DataFrame, featuresCol: String, labelCol: String): DataFrame = { val spark = dataset.sparkSession import spark.implicits._ SchemaUtils.checkColumnType(dataset.schema, featuresCol, new VectorUDT) SchemaUtils.checkNumericType(dataset.schema, labelCol) val rdd = dataset.select(col(labelCol).cast("double"), col(featuresCol)).as[(Double, Vector)] .rdd.map { case (label, features) => OldLabeledPoint(label, OldVectors.fromML(features)) } val testResults = OldStatistics.chiSqTest(rdd) val pValues: Vector = Vectors.dense(testResults.map(_.pValue)) val degreesOfFreedom: Array[Int] = testResults.map(_.degreesOfFreedom) val statistics: Vector = Vectors.dense(testResults.map(_.statistic)) spark.createDataFrame(Seq(ChiSquareResult(pValues, degreesOfFreedom, statistics))) } }
4.执行结果
三.总结器
1.简介
其提供矢量列汇总统计DataFrame的Summarizer。可以度量按列的最大值、最小值、平均值、方差和非零个数,以及总数。
2.代码实现
1 package ml 2 3 import org.apache.log4j.{Level, Logger} 4 import org.apache.spark.ml.linalg.Vectors 5 import org.apache.spark.ml.stat.Summarizer._ // 导入总结器 6 import org.apache.spark.sql.SparkSession 7 import org.apache.spark.ml.linalg.Vector 8 9 /** 10 * Created by Administrator on 2019/11/28. 11 */ 12 object Summary { 13 Logger.getLogger("org").setLevel(Level.WARN) 14 def main(args: Array[String]) { 15 val spark = SparkSession.builder().appName(s"${this.getClass.getSimpleName}").master("local[2]").getOrCreate() 16 import spark.implicits._// 导入,否则无法使用toDF算子 17 18 /** 19 * features数据个数不一致时报错: 20 * Dimensions mismatch when merging with another summarizer. Expecting 3 but got 2. 21 */ 22 val data = Seq( 23 (Vectors.dense(2.0, 3.0, 5.0), 1.0), 24 (Vectors.dense(4.0, 6.0, 8.0), 2.0) 25 ) 26 27 val df = data.toDF("features", "weight") 28 29 /** 30 * 计算均值时考虑权重 31 * [(2.0*1+4.0*2)/3,(3.0*1+6.0*2)/3,(5.0*1+8.0*2)/3) = [3.333333333333333,5.0,7.0] 32 * 方差的计算不考虑权重 33 */ 34 val (meanVal, varianceVal) = df.select(metrics("mean", "variance").summary($"features", $"weight").as("summary")) 35 .select("summary.mean", "summary.variance") 36 .as[(Vector, Vector)].first() 37 38 println(s"with weight:mean = ${meanVal},variance = ${varianceVal}") 39 40 /** 41 * 计算均值,无权重 42 * [(2.0+4.0)/2,(3.0+6.0)/2,(5.0+8.0)/2) = [3.0,4.5,6.5] 43 */ 44 val (meanVal2, varianceVal2) = df.select(mean($"features"), variance($"features")) 45 .as[(Vector, Vector)].first() 46 47 println(s"with weight:mean = ${meanVal2}, variance = ${varianceVal2}") 48 } 49 }
3.源码分析
/** * Tools for vectorized statistics on MLlib Vectors. * * The methods in this package provide various statistics for Vectors contained inside DataFrames. * * This class lets users pick the statistics they would like to extract for a given column. Here is * an example in Scala: * {{{ * import org.apache.spark.ml.linalg._ * import org.apache.spark.sql.Row * val dataframe = ... // Some dataframe containing a feature column and a weight column * val multiStatsDF = dataframe.select( * Summarizer.metrics("min", "max", "count").summary($"features", $"weight") * val Row(Row(minVec, maxVec, count)) = multiStatsDF.first() * }}} * * If one wants to get a single metric, shortcuts are also available: * {{{ * val meanDF = dataframe.select(Summarizer.mean($"features")) * val Row(meanVec) = meanDF.first() * }}} * * Note: Currently, the performance of this interface is about 2x~3x slower than using the RDD * interface. */ @Experimental @Since("2.3.0") object Summarizer extends Logging { import SummaryBuilderImpl._ /** * Given a list of metrics, provides a builder that it turns computes metrics from a column. * * See the documentation of [[Summarizer]] for an example. * * The following metrics are accepted (case sensitive): * - mean: a vector that contains the coefficient-wise mean. * - variance: a vector tha contains the coefficient-wise variance. * - count: the count of all vectors seen. * - numNonzeros: a vector with the number of non-zeros for each coefficients * - max: the maximum for each coefficient. * - min: the minimum for each coefficient. * - normL2: the Euclidean norm for each coefficient. * - normL1: the L1 norm of each coefficient (sum of the absolute values). * @param metrics metrics that can be provided. * @return a builder. * @throws IllegalArgumentException if one of the metric names is not understood. * * Note: Currently, the performance of this interface is about 2x~3x slower then using the RDD * interface. */ @Since("2.3.0") @scala.annotation.varargs def metrics(metrics: String*): SummaryBuilder = { require(metrics.size >= 1, "Should include at least one metric") val (typedMetrics, computeMetrics) = getRelevantMetrics(metrics) new SummaryBuilderImpl(typedMetrics, computeMetrics) } @Since("2.3.0") def mean(col: Column, weightCol: Column): Column = { getSingleMetric(col, weightCol, "mean") } @Since("2.3.0") def mean(col: Column): Column = mean(col, lit(1.0)) @Since("2.3.0") def variance(col: Column, weightCol: Column): Column = { getSingleMetric(col, weightCol, "variance") } @Since("2.3.0") def variance(col: Column): Column = variance(col, lit(1.0)) ...
4.执行结果