- c++多态 详解
蠢 愚
c++c++开发语言
0.多态的概念:多态的概念:相同的消息可能会送给多个不同的类别之对象,而系统可依据对象所属类别,引发对应类别的方法,而有不同的行为。简单来说,所谓多态意指相同的消息给予不同的对象会引发不同的动作。多态分为静态多态与动态多态。静态与动态是针对编译期间与运行期间而言的。静态多态是编译期间就确定要调用什么了,比如函数重载,底层是将函数名与参数按照规则重新命名动态多态是运行期间才能知道调用什么,下文主要讲
- 【详解】数据库E-R图——医院计算机管理系统
嵌入式crafter
数据库
题目某医院病房计算机管理中需要如下信息:科室:科室名,科室地址,科室电话,医生姓名病房:病房号,床位号,所属科室名医生:工作证号,姓名,性别,出生日期,联系电话,职称,所属科室名病人:病历号,姓名,性别,诊断记录,主管医生,病房号其中,一个科室有多个病房、多个医生,一个病房只能属于一个科室,一个医生只属于一个科室,但可负责多个病人的诊治,一个病人的主管医生只有一个。完成如下设计:(1)设计该计算机
- 详解贪心算法
凭君语未可
算法软考算法贪心算法
贪心算法什么是贪心算法?贪心算法的特点贪心算法的应用场景贪心算法的基本思路贪心算法的经典应用1.活动选择问题2.最小硬币找零问题3.霍夫曼编码问题贪心算法的正确性贪心算法的优缺点总结什么是贪心算法?贪心算法(GreedyAlgorithm)是一种基于每一步都选择当前最优解的算法设计思想。它在每个阶段总是做出在当前看来最优的选择(局部最优解),而不回溯或考虑整个问题的全局最优性。它期望通过这样逐步构
- 一张图详解开源监控夜莺(Nightingale)的架构
夜莺开源监控
开源架构夜莺监控Nightingale开源夜莺
夜莺监控是一款开源云原生观测分析工具,采用All-in-One的设计理念,集数据采集、可视化、监控告警、数据分析于一体,与云原生生态紧密集成,提供开箱即用的企业级监控分析和告警能力。夜莺于2020年3月20日,在github上发布v1版本,已累计迭代100多个版本。夜莺最初由滴滴开发和开源,并于2022年5月11日,捐赠予中国计算机学会开源发展委员会(CCFODC),为CCFODC成立后接受捐赠的
- 连通无向图一般中心的算法及其matlab程序详解
夏天天天天天天天#
图论算法matlab图论
#################本文为学习《图论算法及其MATLAB实现》的学习笔记#################若服务点只允许取在各顶点上,而服务对象却取在各顶点及各边(或弧)上的点,则在所有顶点中选定一个顶点作为图的一般中心其条件是该点离它本身的最远服务对象(包括顶点及各边(或弧)上的点)的距离达到极小值。寻找无向图的一般中心对解决网络最佳服务点确定的问题是十分有效的,使得服务对象的范围
- Linux三剑客之grep命令详解
promise524
Linuxlinux服务器pythonshellbash后端运维
grep是Linux中最常用的文本搜索工具,用于在文件或文本输出中查找与指定模式匹配的行。它支持基本正则表达式、扩展正则表达式、多文件搜索、递归搜索等多种功能,非常适合过滤、搜索和提取文本内容。1.grep的基本语法grep[选项]模式[文件...]模式:搜索的文本模式,可以是普通字符串或正则表达式。[文件...]:要搜索的文件。如果没有指定文件,grep会从标准输入中读取数据。2.常用选项-i:
- Linux tar.gz、tar、bz2、zip 等解压缩、压缩命令详解
虫儿飞..
LINUX操作系统linux运维服务器
tar最常用的打包命令是tar,使用tar程序打出来的包我们常称为tar包,tar包文件的命令通常都是以.tar结尾的。生成tar包后,就可以用其它的程序来进行压缩了,所以首先就来讲讲tar命令的基本用法。tar命令的选项有很多(用mantar可以查看到),但常用的就那么几个选项,下面来举例说明一下:#tar-cfall.tar*.jpg这条命令是将所有.jpg的文件打成一个名为all.tar的包
- 公众号开通流量主步骤详解:如何轻松实现广告变现?
优惠券高省
在微信公众号运营的过程中,许多运营者都希望通过各种方式实现变现,其中,开通流量主功能成为了一个备受关注的途径。那么,公众号如何开通流量主呢?本文将为您详细解析公众号开通流量主的步骤,帮助您轻松实现广告变现。3个怪异的现象,正在冲击"一夫一妻制",动摇家庭根基!未来这3种婚姻模式,可能“取代”传统婚姻制度《人民日报》金句:"万般皆苦,唯有自渡,活着就要遇山开山,见水架桥,其实一直陪着你的,永远都是那
- 【java】【springboot】启动方法注解详解
纳米小川
javaspringbootmybatis
在SpringBoot中,启动方法通常是main方法,它位于应用程序的主类中,负责启动Spring应用上下文。一、@SpringBootApplication@SpringBootApplication是SpringBoot中一个非常核心的注解,它是一个复合注解,包含了三个关键的注解:@SpringBootConfiguration、@EnableAutoConfiguration和@Compon
- 即时通讯开发之TCP/IP中的TCP 协议概述
wecloud1314
tcp/ip网络udp
终于看到了TCP协议,这是TCP/IP详解里面最重要也是最精彩的部分,要花大力气来读。前面的TFTP和BOOTP都是一些简单的协议,就不写笔记了,写起来也没啥东西。TCP和UDP处在同一层---运输层,但是TCP和UDP最不同的地方是,TCP提供了一种可靠的数据传输服务,TCP是面向连接的,也就是说,利用TCP通信的两台主机首先要经历一个“拨打电话”的过程,等到通信准备结束才开始传输数据,最后结束
- 美团app优惠券怎么使用 美团使用优惠券订餐方法【详解】
高省_飞智666600
掌握美团优惠券的使用技巧,就如同掌握了一门艺术。每一款优惠券都有其独特的魅力,而你的任务,就是细心领悟,找到最能触动你心弦的优惠券,让它为你的生活增添一份惊喜。【高省】APP(高佣金领导者)是一个自用省钱佣金高,分享推广赚钱多的平台,百度有几百万篇报道,运行三年,稳定可靠。高省APP,是2021年推出的平台,0投资,0风险、高省APP佣金更高,模式更好,终端用户不流失。高省是公认的返利最高的软件。
- 详解TCP的三次握手
汪先声
tcp/ip网络协议网络
TCP(三次握手)是指在建立一个可靠的传输控制协议(TCP)连接时,客户端和服务器之间的三步交互过程。这个过程的主要目的是确保连接是可靠的、双方的发送与接收能力是正常的,并且可以开始数据传输。下面是对每个步骤的详细解释:1.第一次握手:客户端发送SYN过程:客户端(A)向服务器(B)发送一个同步报文段(SYN,SynchronizeSequenceNumber),表示它想要与服务器建立连接。目的:
- 微信小程序中的实时通讯:TCP/UDP 协议实现详解
人工智能的苟富贵
前端小程序微信小程序tcp/ipudp
文章目录前言一、实时通讯的基础知识二、微信小程序中TCP/UDP的支持2.1TCP实现2.2UDP实现三、实现即时通讯的基本架构四、实际开发中的注意事项4.1网络环境问题4.2数据格式与协议设计4.3消息重发机制五、实时通讯中的性能优化5.1减少不必要的通信5.2数据压缩5.3异步通信与心跳机制六、使用场景七、总结前言在现代应用程序中,实时通讯已成为用户体验的关键组成部分。无论是在线聊天、游戏、还
- Dubbo 与 Zookeeper 在项目中的应用:原理与实现详解
CopyLower
学习Javadubbozookeeper分布式
引言在微服务架构日益普及的今天,如何实现服务的高效调用和管理成为了关键问题。Dubbo作为阿里巴巴开源的高性能RPC框架,在分布式服务治理方面具有显著的优势。Zookeeper作为一款分布式协调服务,能够高效地管理和协调服务节点信息。因此,Dubbo与Zookeeper的结合不仅能够提供服务注册与发现机制,还能实现更高效的服务治理。在本文中,我们将深入探讨Dubbo和Zookeeper的原理、如何
- 隔离上网技术详解:打造安全隔离的网络环境
cnsinda_htt
源代码防泄密源代码防泄漏源代码加密安全
SDC沙盒数据防泄密系统(安全上网,隔离上网)•深信达SDC沙盒数据防泄密系统,是专门针对敏感数据进行防泄密保护的系统,根据隔离上网和安全上网的原则实现数据的代码级保护,不会影响工作效率,不影响正常使用。所有敏感信息和文件都自动加密,从而得到有效的范围控制,防止泄密。沙盒数据防泄密系统,系统架构如下:•管理端:系统控制中心,策略管理•机密端:源代码及设计文档版本管理服务器,可以有复数台•外发审核服
- C生万物 函数的讲解与剖析【内附众多案例详解】
2401_84170337
c语言java开发语言
printf("交换前:a=%d,b=%d\n",a,b);swap(a,b);printf("交换后:a=%d,b=%d\n",a,b);return0;}*来看一下运行结果。可以看到两个数并没有发生交换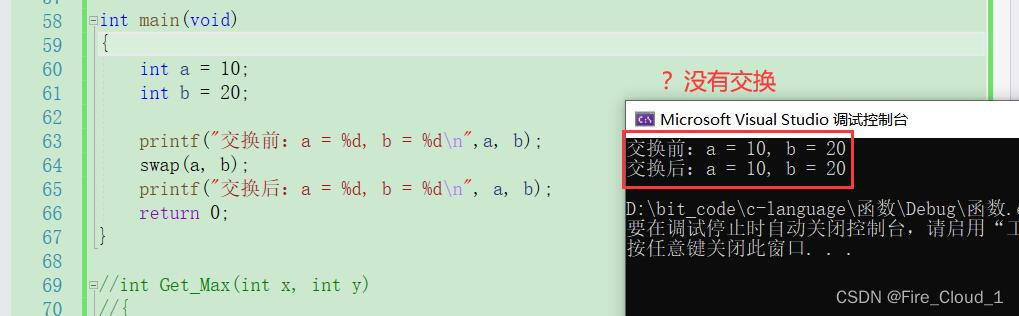*那有小伙伴就
- mysql myisam 默认隔离级别_MySQL事务隔离级别详解
weixin_39656513
mysqlmyisam默认隔离级别
前两天面试,问到了四种隔离级别,当时觉得大多数数据库都为readcommitted,结果没想到mysql是个例外。在此做一下隔离级别和各种数据库锁的使用。首先说一下ACID四大特性:四大特性·原子性事务必须是原子工作单元;对于其数据修改,要么全都执行,要么全都不执行。通常,与某个事务关联的操作具有共同的目标,并且是相互依赖的。如果系统只执行这些操作的一个子集,则可能会破坏事务的总体目标。原子性消除
- 【百日算法计划】:每日一题,见证成长(013)
码上一元
数据结构与算法算法
题目回文链表给你一个单链表的头节点head,请你判断该链表是否为回文链表。如果是,返回true;否则,返回false。输入:head=[1,2,2,1]输出:true思路找到中间节点反转后半部分链表前后链表顺序比对publicbooleanisPalindrome2(ListNodehead){if(head==null||head.next==null)returntrue;ListNodep=
- 2018农信社备考知识点精选:多定义判断
云南华图金领人
农信社笔试中判断推理-定义判断-多定义判断怎么击破,华图农村信用社招聘网为你解答。A.如果我发挥得稍微差一点,就与奖牌失之交臂了知识点:判断推理-定义判断-多定义判断知识点详解:多定义判断的做题方法:在题干定位所提问的定义,再运用单定义判断的方法,寻找要件,提取关键信息,对比选项。要件主体:定义的发出者,是实践活动和认识活动的承担者,与定义相符的选项的主体应该与定义的主体一致。客体:主体行为所指向
- C++ 判断语句详解
chengong9988
编程c++开发语言
C++判断语句详解在C++编程中,判断语句(ConditionalStatements)是一种用于根据条件执行不同代码块的控制结构。它们使得程序可以根据特定的条件来做出决策,从而实现更灵活和智能的行为。本文将介绍C++中常见的判断语句,包括if、if-else、if-elseif-else和switch,并给出详细的示例帮助您理解和应用这些语句。1.if语句if语句用于在条件为真时执行特定的代码块
- Unity3D DOTS系列之Struct Change核心机制分析详解
Thomas_YXQ
开发语言Unity3D游戏unity架构
引言Unity3D的DOTS(Data-OrientedTechnologyStack)体系为游戏开发带来了革命性的变化,它通过ECS(EntityComponentSystem)模型,将游戏中的对象(Entity)、属性(Component)和行为(System)分离,以数据驱动的方式来提高游戏的性能和可扩展性。在DOTS体系中,StructChange是一个核心的内存管理机制,它涉及对Enti
- Unity3D GPUDriven渲染详解
Thomas_YXQ
开发语言Unity3D架构游戏Unity
前言Unity3D中的GPUDriven渲染技术是一种通过最大化GPU的利用,减少CPU负担,从而提高渲染效率和帧率的方法。其核心思想是将更多的渲染任务转移到GPU上,充分利用现代图形硬件(显卡)的性能。以下是该技术的几个关键组件和它们的作用:对惹,这里有一个游戏开发交流小组,大家可以点击进来一起交流一下开发经验呀!1.BatchRendererGroup(BRG)BRG是Unity中用于批处理渲
- Unity3D帧同步模式的网络游戏详解
Thomas_YXQ
游戏开发Unity3DUnity开发语言ui
帧同步概述帧同步(FrameSynchronization)是指在网络游戏中,多个客户端在同一时刻执行相同的游戏逻辑,确保各个客户端的游戏状态保持一致。这种同步方式对于实现公平的多人游戏和减少网络延迟对游戏体验的影响至关重要。Unity3D作为一款强大的跨平台游戏引擎,提供了丰富的工具和接口来实现帧同步。对惹,这里有一个游戏开发交流小组,大家可以点击进来一起交流一下开发经验呀!实现步骤1.确定帧率
- C# WPF中的GUI多线程技巧详解
zls365365
c#wpf开发语言
1.使用BackgroundWorker组件代码示例:publicpartialclassMainWindow:Window{privateBackgroundWorkerbackgroundWorker=newBackgroundWorker();publicMainWindow(){InitializeComponent();backgroundWorker.DoWork+=Backgroun
- Python浏览器指纹反爬详解(包含案例)——blog10
总得跑一个
python网络爬虫selenium
目录概述案例实操目标分析补充开始由此可以得到方法一:直接从api拿数据方法二:伪装selenium.webdriver测试测试用HTML如下:爬取失败——分析与思考改进最后附上使用selenium破解目标网站浏览器指纹的完整代码:觉得有帮助的小伙伴还请点个关注概述浏览器指纹是由浏览器类型、版本号、操作系统、屏幕分辨率、时区、插件、字体等信息组合而成的唯一标识,可以用于区分不同的用户。通过比对请求中
- java class 获取类_Java中通过Class类获取Class对象的方法详解
洪九(李戈)
javaclass获取类
Java中通过Class类获取Class对象的方法详解前言本文主要给大家介绍的是关于Java通过Class类获取Class对象的相关内容,分享出来供大家参考学习,下面话不多说了,来一起看看详细的介绍:阅读API的Class类得知,Class没有公共构造方法。Class对象是在加载类时由Java虚拟机以及通过调用类加载器中的defineClass方法自动构造的获取Class对象的三种方式(实例采用P
- C++线程、多线程教程详解(全网最全、示例最多、最详细)(第一篇)
shuai_258
c++c++全套攻略c++多线程c++
目录A、线程/多线程基础一、C++11创建线程的几种方式1.1使用函数指针1.2使用lambda表达式1.3使用成员函数1.4使用可调用对象(Functor)二、定义一个线程类三、join()与detach()的详细用法及区别3.1join()的用法3.2detach()的用法3.3join()与detach()的区别总结四、std::this_thread4.1、主要功能std::this_th
- 服务器的种类详解,让个人站长了解服务器
安全过了马路却撞了大树
服务器英文名称为“Server”,指的是网络环境下为客户机(Client)提供某种服务的专用计算机,服务器安装有网络操作系统(如Windows2000Server、Linux、Unix等)和各种服务器应用系统软件(如Web服务、电子邮件服务)的计算机。这里的“客户机”指安装有DOS、Windows9x等普通用户使用的操作系统的计算机。服务器的处理速度和系统可靠性都要比普通PC要高得多,因为服务器是
- Java 正则表达式详解
艾伦~耶格尔
Java初级java正则表达式开发语言学习
正则表达式(RegularExpression,简称regex)是一种强大的文本处理工具,可以用来匹配、搜索和替换文本中的特定模式。在Java中,正则表达式由java.util.regex包提供支持。1.理解正则表达式语法正则表达式使用特殊的字符和符号来定义匹配模式。一些常用的元字符如下:.:匹配任意单个字符*:匹配前面的字符零次或多次+:匹配前面的字符一次或多次?:匹配前面的字符零次或一次[]:
- golang学习笔记12——Go 语言内存管理详解
GoppViper
golang学习笔记golang学习笔记编程语言golang内存管理内存优化后端
推荐学习文档golang应用级os框架,欢迎star基于golang开发的一款超有个性的旅游计划app经历golang实战大纲golang优秀开发常用开源库汇总golang学习笔记01——基本数据类型golang学习笔记02——gin框架及基本原理golang学习笔记03——gin框架的核心数据结构golang学习笔记04——如何真正写好Golang代码?golang学习笔记05——golang协
- java类加载顺序
3213213333332132
java
package com.demo;
/**
* @Description 类加载顺序
* @author FuJianyong
* 2015-2-6上午11:21:37
*/
public class ClassLoaderSequence {
String s1 = "成员属性";
static String s2 = "
- Hibernate与mybitas的比较
BlueSkator
sqlHibernate框架ibatisorm
第一章 Hibernate与MyBatis
Hibernate 是当前最流行的O/R mapping框架,它出身于sf.net,现在已经成为Jboss的一部分。 Mybatis 是另外一种优秀的O/R mapping框架。目前属于apache的一个子项目。
MyBatis 参考资料官网:http:
- php多维数组排序以及实际工作中的应用
dcj3sjt126com
PHPusortuasort
自定义排序函数返回false或负数意味着第一个参数应该排在第二个参数的前面, 正数或true反之, 0相等usort不保存键名uasort 键名会保存下来uksort 排序是对键名进行的
<!doctype html>
<html lang="en">
<head>
<meta charset="utf-8&q
- DOM改变字体大小
周华华
前端
<!DOCTYPE html PUBLIC "-//W3C//DTD XHTML 1.0 Transitional//EN" "http://www.w3.org/TR/xhtml1/DTD/xhtml1-transitional.dtd">
<html xmlns="http://www.w3.org/1999/xhtml&q
- c3p0的配置
g21121
c3p0
c3p0是一个开源的JDBC连接池,它实现了数据源和JNDI绑定,支持JDBC3规范和JDBC2的标准扩展。c3p0的下载地址是:http://sourceforge.net/projects/c3p0/这里可以下载到c3p0最新版本。
以在spring中配置dataSource为例:
<!-- spring加载资源文件 -->
<bean name="prope
- Java获取工程路径的几种方法
510888780
java
第一种:
File f = new File(this.getClass().getResource("/").getPath());
System.out.println(f);
结果:
C:\Documents%20and%20Settings\Administrator\workspace\projectName\bin
获取当前类的所在工程路径;
如果不加“
- 在类Unix系统下实现SSH免密码登录服务器
Harry642
免密ssh
1.客户机
(1)执行ssh-keygen -t rsa -C "
[email protected]"生成公钥,xxx为自定义大email地址
(2)执行scp ~/.ssh/id_rsa.pub root@xxxxxxxxx:/tmp将公钥拷贝到服务器上,xxx为服务器地址
(3)执行cat
- Java新手入门的30个基本概念一
aijuans
javajava 入门新手
在我们学习Java的过程中,掌握其中的基本概念对我们的学习无论是J2SE,J2EE,J2ME都是很重要的,J2SE是Java的基础,所以有必要对其中的基本概念做以归纳,以便大家在以后的学习过程中更好的理解java的精髓,在此我总结了30条基本的概念。 Java概述: 目前Java主要应用于中间件的开发(middleware)---处理客户机于服务器之间的通信技术,早期的实践证明,Java不适合
- Memcached for windows 简单介绍
antlove
javaWebwindowscachememcached
1. 安装memcached server
a. 下载memcached-1.2.6-win32-bin.zip
b. 解压缩,dos 窗口切换到 memcached.exe所在目录,运行memcached.exe -d install
c.启动memcached Server,直接在dos窗口键入 net start "memcached Server&quo
- 数据库对象的视图和索引
百合不是茶
索引oeacle数据库视图
视图
视图是从一个表或视图导出的表,也可以是从多个表或视图导出的表。视图是一个虚表,数据库不对视图所对应的数据进行实际存储,只存储视图的定义,对视图的数据进行操作时,只能将字段定义为视图,不能将具体的数据定义为视图
为什么oracle需要视图;
&
- Mockito(一) --入门篇
bijian1013
持续集成mockito单元测试
Mockito是一个针对Java的mocking框架,它与EasyMock和jMock很相似,但是通过在执行后校验什么已经被调用,它消除了对期望 行为(expectations)的需要。其它的mocking库需要你在执行前记录期望行为(expectations),而这导致了丑陋的初始化代码。
&nb
- 精通Oracle10编程SQL(5)SQL函数
bijian1013
oracle数据库plsql
/*
* SQL函数
*/
--数字函数
--ABS(n):返回数字n的绝对值
declare
v_abs number(6,2);
begin
v_abs:=abs(&no);
dbms_output.put_line('绝对值:'||v_abs);
end;
--ACOS(n):返回数字n的反余弦值,输入值的范围是-1~1,输出值的单位为弧度
- 【Log4j一】Log4j总体介绍
bit1129
log4j
Log4j组件:Logger、Appender、Layout
Log4j核心包含三个组件:logger、appender和layout。这三个组件协作提供日志功能:
日志的输出目标
日志的输出格式
日志的输出级别(是否抑制日志的输出)
logger继承特性
A logger is said to be an ancestor of anothe
- Java IO笔记
白糖_
java
public static void main(String[] args) throws IOException {
//输入流
InputStream in = Test.class.getResourceAsStream("/test");
InputStreamReader isr = new InputStreamReader(in);
Bu
- Docker 监控
ronin47
docker监控
目前项目内部署了docker,于是涉及到关于监控的事情,参考一些经典实例以及一些自己的想法,总结一下思路。 1、关于监控的内容 监控宿主机本身
监控宿主机本身还是比较简单的,同其他服务器监控类似,对cpu、network、io、disk等做通用的检查,这里不再细说。
额外的,因为是docker的
- java-顺时针打印图形
bylijinnan
java
一个画图程序 要求打印出:
1.int i=5;
2.1 2 3 4 5
3.16 17 18 19 6
4.15 24 25 20 7
5.14 23 22 21 8
6.13 12 11 10 9
7.
8.int i=6
9.1 2 3 4 5 6
10.20 21 22 23 24 7
11.19
- 关于iReport汉化版强制使用英文的配置方法
Kai_Ge
iReport汉化英文版
对于那些具有强迫症的工程师来说,软件汉化固然好用,但是汉化不完整却极为头疼,本方法针对iReport汉化不完整的情况,强制使用英文版,方法如下:
在 iReport 安装路径下的 etc/ireport.conf 里增加红色部分启动参数,即可变为英文版。
# ${HOME} will be replaced by user home directory accordin
- [并行计算]论宇宙的可计算性
comsci
并行计算
现在我们知道,一个涡旋系统具有并行计算能力.按照自然运动理论,这个系统也同时具有存储能力,同时具备计算和存储能力的系统,在某种条件下一般都会产生意识......
那么,这种概念让我们推论出一个结论
&nb
- 用OpenGL实现无限循环的coverflow
dai_lm
androidcoverflow
网上找了很久,都是用Gallery实现的,效果不是很满意,结果发现这个用OpenGL实现的,稍微修改了一下源码,实现了无限循环功能
源码地址:
https://github.com/jackfengji/glcoverflow
public class CoverFlowOpenGL extends GLSurfaceView implements
GLSurfaceV
- JAVA数据计算的几个解决方案1
datamachine
javaHibernate计算
老大丢过来的软件跑了10天,摸到点门道,正好跟以前攒的私房有关联,整理存档。
-----------------------------华丽的分割线-------------------------------------
数据计算层是指介于数据存储和应用程序之间,负责计算数据存储层的数据,并将计算结果返回应用程序的层次。J
&nbs
- 简单的用户授权系统,利用给user表添加一个字段标识管理员的方式
dcj3sjt126com
yii
怎么创建一个简单的(非 RBAC)用户授权系统
通过查看论坛,我发现这是一个常见的问题,所以我决定写这篇文章。
本文只包括授权系统.假设你已经知道怎么创建身份验证系统(登录)。 数据库
首先在 user 表创建一个新的字段(integer 类型),字段名 'accessLevel',它定义了用户的访问权限 扩展 CWebUser 类
在配置文件(一般为 protecte
- 未选之路
dcj3sjt126com
诗
作者:罗伯特*费罗斯特
黄色的树林里分出两条路,
可惜我不能同时去涉足,
我在那路口久久伫立,
我向着一条路极目望去,
直到它消失在丛林深处.
但我却选了另外一条路,
它荒草萋萋,十分幽寂;
显得更诱人,更美丽,
虽然在这两条小路上,
都很少留下旅人的足迹.
那天清晨落叶满地,
两条路都未见脚印痕迹.
呵,留下一条路等改日再
- Java处理15位身份证变18位
蕃薯耀
18位身份证变15位15位身份证变18位身份证转换
15位身份证变18位,18位身份证变15位
>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>>
蕃薯耀 201
- SpringMVC4零配置--应用上下文配置【AppConfig】
hanqunfeng
springmvc4
从spring3.0开始,Spring将JavaConfig整合到核心模块,普通的POJO只需要标注@Configuration注解,就可以成为spring配置类,并通过在方法上标注@Bean注解的方式注入bean。
Xml配置和Java类配置对比如下:
applicationContext-AppConfig.xml
<!-- 激活自动代理功能 参看:
- Android中webview跟JAVASCRIPT中的交互
jackyrong
JavaScripthtmlandroid脚本
在android的应用程序中,可以直接调用webview中的javascript代码,而webview中的javascript代码,也可以去调用ANDROID应用程序(也就是JAVA部分的代码).下面举例说明之:
1 JAVASCRIPT脚本调用android程序
要在webview中,调用addJavascriptInterface(OBJ,int
- 8个最佳Web开发资源推荐
lampcy
编程Web程序员
Web开发对程序员来说是一项较为复杂的工作,程序员需要快速地满足用户需求。如今很多的在线资源可以给程序员提供帮助,比如指导手册、在线课程和一些参考资料,而且这些资源基本都是免费和适合初学者的。无论你是需要选择一门新的编程语言,或是了解最新的标准,还是需要从其他地方找到一些灵感,我们这里为你整理了一些很好的Web开发资源,帮助你更成功地进行Web开发。
这里列出10个最佳Web开发资源,它们都是受
- 架构师之面试------jdk的hashMap实现
nannan408
HashMap
1.前言。
如题。
2.详述。
(1)hashMap算法就是数组链表。数组存放的元素是键值对。jdk通过移位算法(其实也就是简单的加乘算法),如下代码来生成数组下标(生成后indexFor一下就成下标了)。
static int hash(int h)
{
h ^= (h >>> 20) ^ (h >>>
- html禁止清除input文本输入缓存
Rainbow702
html缓存input输入框change
多数浏览器默认会缓存input的值,只有使用ctl+F5强制刷新的才可以清除缓存记录。
如果不想让浏览器缓存input的值,有2种方法:
方法一: 在不想使用缓存的input中添加 autocomplete="off";
<input type="text" autocomplete="off" n
- POJO和JavaBean的区别和联系
tjmljw
POJOjava beans
POJO 和JavaBean是我们常见的两个关键字,一般容易混淆,POJO全称是Plain Ordinary Java Object / Pure Old Java Object,中文可以翻译成:普通Java类,具有一部分getter/setter方法的那种类就可以称作POJO,但是JavaBean则比 POJO复杂很多, Java Bean 是可复用的组件,对 Java Bean 并没有严格的规
- java中单例的五种写法
liuxiaoling
java单例
/**
* 单例模式的五种写法:
* 1、懒汉
* 2、恶汉
* 3、静态内部类
* 4、枚举
* 5、双重校验锁
*/
/**
* 五、 双重校验锁,在当前的内存模型中无效
*/
class LockSingleton
{
private volatile static LockSingleton singleton;
pri