[论文翻译]UNet++: A Nested U-Net Architecture for Medical Image Segmentation
UNet++论文: 地址
UNet++: A Nested U-Net Architecture for Medical Image Segmentation
UNet++:一个用于医学图像分割的嵌套的U-Net网络结构
Zongwei Zhou, Md Mahfuzur Rahman Siddiquee,Nima Tajbakhsh, and Jianming Liang
周纵苇等人
Arizona State University
亚利桑那州立大学
Abstract 摘要
In this paper, we present UNet++, a new, more powerful architecture for medical image segmentation. Our architecture is essentially a deeply-supervised encoder-decoder network where the encoder and decoder sub-networks are connected through a series of nested, dense skip pathways. The re-designed skip pathways aim at reducing the semantic gap between the feature maps of the encoder and decoder sub-networks. We argue that the optimizer would deal with an easier learning task when the feature maps from the decoder and encoder networks are semantically similar. We have evaluated UNet++ in comparison with U-Net and wide U-Net architectures across multiple medical image segmentation tasks: nodule segmentation in the low-dose CT scans of chest, nuclei segmentation in the microscopy images, liver segmentation in abdominal CT scans, and polyp segmentation in colonoscopy videos. Our experiments demonstrate that UNet++ with deep supervision achieves an average IoU gain of 3.9 and 3.4 points over U-Net and wide U-Net, respectively
在这一篇论文里, 我们展示了一个新的且更强的网络结构UNet++用于医学图像分割.我们的结构本质上是一个深度监督(deeply-supervised)的编码-解码网络,编码和解码的子网络通过一个系列嵌套的稠密的跳跃路径来相互连接.这种重新设计的跳跃连接主要是用来降低编码解码子网络中特征图的语义缺失.当编码解码网络的特征图语义类似时,我们认为这种优化器可以解决一个更简单的学习任务.我们在多种医学图像分割任务(multiple medical image segmentation tasks)中比较了UNet++、UNet以及UNet衍生网络的预测效果:小剂量的CT胸透扫描瘤分割、显微镜里的细胞核分割、腹部CT的肝脏分割以及结肠息肉分割.我们的实验说明了具有深度监督结构的UNet++取得平均3.9的IoU和3.4 points 分别超过了U-Net和U-Net的衍生.
1 Introduction 介绍
The state-of-the-art models for image segmentation are variants of the encoderdecoder architecture like U-Net [9] and fully convolutional network (FCN) [8]. These encoder-decoder networks used for segmentation share a key similarity: skip connections, which combine deep, semantic, coarse-grained feature maps from the decoder sub-network with shallow, low-level, fine-grained feature maps from the encoder sub-network. The skip connections have proved effective in recovering fine-grained details of the target objects; generating segmentation masks with fine details even on complex background. Skip connections is also fundamental to the success of instance-level segmentation models such as MaskRCNN, which enables the segmentation of occluded objects. Arguably, image segmentation in natural images has reached a satisfactory level of performance, but do these models meet the strict segmentation requirements of medical images?
图像分割的SOTA模型有各种基于像U-Net和FCN解码编码结构的变体.这些被用来分割的解码-编码网络结构都给我们展示了一个关键且相似的东西:即跳跃连接,用来结合来自解码子网络中深度语义粗略的特征图和来自编码子网络中浅层低级详细的特征图.这种跳跃连接已经证明在恢复目标对象的详细细节上是有效的;可以最终在复杂的背景中产生更加细节的分割图.跳跃连接也是一个用来实现实例分割 (instance-level)模型的基本组成要素,例如MaskRCNN能够识别并分割遮挡的对象.可以证明, 在自然图像中的图像分割已经达到了满意的表现,但是用这些模型是否可以满足医学图像中的严格的分割要求呢?
Segmenting lesions or abnormalities in medical images demands a higher level of accuracy than what is desired in natural images. While a precise segmentation mask may not be critical in natural images, even marginal segmentation errors in medical images can lead to poor user experience in clinical settings. For instance,the subtle spiculation patterns around a nodule may indicate nodule malignancy; and therefore, their exclusion from the segmentation masks would lower the credibility of the model from the clinical perspective. Furthermore, inaccurate segmentation may also lead to a major change in the subsequent computer-generated diagnosis. For example, an erroneous measurement of nodule growth in longitudinal studies can result in the assignment of an incorrect Lung-RADS category to a screening patient. It is therefore desired to devise more effective image segmentation architectures that can effectively recover the fine details of the target objects in medical images.
在医学图像中分割病灶(lesions)或者畸形部分需要一个更高级别的分割精度度(accuracy)比自然图像.自然图像中一个精确的分割可能没有那么重要,而在医学图像中微不足道的分割差错可以误导经验不足的使用者在临床配置的时候.举个例子,小瘤附近的细微刺状图案(subtle spiculation patterns)可以指出瘤的恶性;因此,在临床方面他们从分割图判断的结论应该降低模型的可信度.而且,在随后计算机生成(computer-generated)的诊断中,不精确的分割也可能导致一个大的改变.例如一个错误的瘤增长测量可以给一个筛选病人最终一个不正确的肺结节(Lung-RADS)分类分配.因此,在医学图像分割中更加需要设计一个网络可以更有效地恢复目标对象的细节.
To address the need for more accurate segmentation in medical images, we present UNet++, a new segmentation architecture based on nested and dense skip connections. The underlying hypothesis behind our architecture is that the model can more effectively capture fine-grained details of the foreground objects when high-resolution feature maps from the encoder network are gradually enriched prior to fusion with the corresponding semantically rich feature maps from the decoder network. We argue that the network would deal with an easier learning task when the feature maps from the decoder and encoder networks are semantically similar. This is in contrast to the plain skip connections commonly used in U-Net, which directly fast-forward high-resolution feature maps from the encoder to the decoder network, resulting in the fusion of semantically dissimilar feature maps. According to our experiments, the suggested architecture is effective, yielding significant performance gain over U-Net and wide U-Net.
为了设法解决这种在医学图像中更加精准的分割需求,我们提出的UNet++,这个网络是基于嵌套的和稠密的跳跃连接来实现的.即当高分辨率特征图从编码网络逐渐地和解码网络中的相应语义的特征图优先进行融合,这个网络它可以更高效地捕获前景对象的深层(fine-grained)细节,.我们提出的理由是当解码和编码的特征图的语义相似进行融合时,它学习的效果会更好,这与UNet的朴素跳跃连接不同,在UNet中高分辨率特征图快速直接地从编码到解码,结果是不相似语义的特征图之间进行融合.根据我们的实验,UNet++网络是有提升的,并且变现的分数超过了U-Net和wide U-Net.
2 Related Work 相关工作
Long et al. [8] first introduced fully convolutional networks (FCN), while UNet was introduced by Ronneberger et al. [9]. They both share a key idea: skip connections. In FCN, up-sampled feature maps are summed with feature maps skipped from the encoder, while U-Net concatenates them and add convolutions and non-linearities between each up-sampling step. The skip connections have shown to help recover the full spatial resolution at the network output, making fully convolutional methods suitable for semantic segmentation. Inspired by DenseNet architecture [5], Li et al. [7] proposed H-denseunet for liver and liver tumor segmentation. In the same spirit, Drozdzalet al. [2] systematically investigated the importance of skip connections, and introduced short skip connections within the encoder. Despite the minor differences between the above architectures, they all tend to fuse semantically dissimilar feature maps from the encoder and decoder sub-networks, which, according to our experiments, can degrade segmentation performance.
Long et al [8]第一次提出FCN,同一年(2015)Ronneberger et al. [9]提出了UNet(稍微晚点).他们都用一个重要的结构:跳跃连接(操作不同).在FCN中,上采样时的特征图是用来自编码的特征图进行像素点的加操作(summation),而U-Net在每个上采样之间有进行维度拼接的叠操作(concatenation)、卷积和非线性激活函数.无论是哪种跳跃连接,都说明了它在网络的输出中可以帮忙恢复丰富的空间分辨率,使得全卷积方法适合语义分割.受到 DenseNet architecture [5]的启发,Li et al. [7] 提出H-denseunet用于肝脏和肝瘤的分割.同样受到启发,Drozdzalet al. [2]系统性地研究分析了跳跃连接的重要性,而且介绍了在编码里的短跳跃连接.尽管以上的结构有一点的不同,但是他们都趋向于结合编码解码子网络中语义不同的特征图,根据我们实验验证,这样的方式降低了分割的表现.
The other two recent related works are GridNet [3] and Mask-RCNN [4].GridNet is an encoder-decoder architecture wherein the feature maps are wired in a grid fashion, generalizing several classical segmentation architectures. GridNet,however, lacks up-sampling layers between skip connections; and thus, it does not represent UNet++. Mask-RCNN is perhaps the most important meta framework for object detection, classification and segmentation. We would like to note that UNet++ can be readily deployed as the backbone architecture in Mask-RCNN by simply replacing the plain skip connections with the suggested nested dense skip pathways. Due to limited space, we were not able to include results of Mask RCNN with UNet++ as the backbone architecture; however, the interested readers can refer to the supplementary material for further details.
最近另外两个相关工作是GridNet [3] 和 Mask-RCNN [4].GridNet是一个编码-解码结构,里面的特征图被线连成网格型,形成了数个类分割结构.然而GridNet在跳跃连接间缺乏上采样层;因此,它不能代表UNet++.Mask-RCNN也许对于目标检测、分类以及分割是最重要的框架.我们想UNet++可以作为骨干框架而很容易被展开 在Mask-RCNN 通过简单地用嵌套稠密的跳跃路径取代朴素跳跃连接.由于空间的限制,我们不能用UNet++作为骨干框架来包含Mask RCNN的结果.然而, 有兴趣看更多细节的读者可以查阅到补充资料.
Fig. 1: (a) UNet++ consists of an encoder and decoder that are connected through a series of nested dense convolutional blocks. The main idea behind UNet++ is to bridge the semantic gap between the feature maps of the encoder and decoder prior to fusion. For example, the semantic gap between (X0;0,X1;3) is bridged using a dense convolution block with three convolution layers. In the graphical abstract, black indicates the original U-Net, green and blue show dense convolution blocks on the skip pathways, and red indicates deep supervision. Red, green, and blue components distinguish UNet++ from U-Net. (b) Detailed analysis of the first skip pathway of UNet++. (c) UNet++ can be pruned at inference time, if trained with deep supervision
图1:(a)UNet++ 包含编码和解码以及一系列将它们连接的嵌套稠密的卷积块.UNet++背后的主要的思路是在融合之前先架起编码解码的特征图之间的语义不同.例如,在(X0,0 X1,3)之间的语义差异是使用一个有3层卷积的稠密卷积块来架起.在图中,黑色表示原始原来的U-Net,绿色和蓝色表示在跳跃连接上的稠密卷积块,红色表示深度监督.红色,绿色和蓝色的成分是UNet++的用来区别于UNet.(b)UNet++跳跃路径的分析细节 (c) 如果Unet++用深度监督来训练的话,它可以在预测的时候被简化
3 Proposed Network Architecture: UNet++ 推荐的神经网络结构
Fig. 1a shows a high-level overview of the suggested architecture. As seen,UNet++ starts with an encoder sub-network or backbone followed by a decoder sub-network. What distinguishes UNet++ from U-Net (the black components in Fig. 1a) is the re-designed skip pathways (shown in green and blue) that connect the two sub-networks and the use of deep supervision (shown red)
Fig. 1a 展示了UNet++的一个总体的概述.我们可以看到,UNet++用一个编码子网络来或者用跟随这一个解码子网络的骨干来开始.UNet++和U-Net的区别(在Fig.1a中的黑色部分)就是重新设计的跳跃路径(绿色和蓝色),它用来连接两个子网络和深度监督的使用部分(红色)
3.1 Re-designed skip pathways
Re-designed skip pathways transform the connectivity of the encoder and decoder sub-networks. In U-Net, the feature maps of the encoder are directly received in the decoder; however, in UNet++, they undergo a dense convolution block whose number of convolution layers depends on the pyramid level. For example, the skip pathway between nodes X0,0 and X1,3 consists of a dense convolution block with three convolution layers where each convolution layer is preceded by a concatenation layer that fuses the output from the previous convolution layer of the same dense block with the corresponding up-sampled output of the lower dense block. Essentially, the dense convolution block brings the semantic level of the encoder feature maps closer to that of the feature maps awaiting in the decoder. The hypothesis is that the optimizer would face an easier optimization problem when the received encoder feature maps and the corresponding decoder feature maps are semantically similar
这种重新设计的跳跃路径改变了编码和解码子网络的连接性.在U-Net中,解码是直接获取编码中的特征图;然而, 在UNet++中,它们经过一个稠密卷积块,这个块的卷积层数量取决于“金字塔”的级别.例如,节点 X0,0 和 节点 X1,3之间的跳跃路径是由一个有3层卷积的稠密卷积块,每一层卷积都领先于一个连接层,这个连接层结合将相同稠密块的前一个卷积层的输出与相应的上采样低稠密块的输出进行融合。本质上,密集的卷积块使得编码器的特征映射的语义级别更接近于解码器中等待的特征映射的语义级别。假设当接收到的编码器特征映射和相应的解码器特征映射在语义上相似时,优化器将面临更容易的优化问题
Formally, we formulate the skip pathway as follows: let xi,j denote the output of node Xi,j where i indexes the down-sampling layer along the encoder and j indexes the convolution layer of the dense block along the skip pathway. The
stack of feature maps represented by xi,j is computed as从公式上表示跳跃路径如下:让xi,j表示节点 Xi,j的输出.i指的是沿着编码方向的哪个下采样层,j指的是沿着跳跃路径方向上稠密块的那个卷积层.这一堆特征图用 xi,j 表示,计算如下:
where function H(·) is a convolution operation followed by an activation function, U(·) denotes an up-sampling layer, and [ ] denotes the concatenation layer.Basically, nodes at level j = 0 receive only one input from the previous layer of the encoder; nodes at level j = 1 receive two inputs, both from the encoder sub-network but at two consecutive levels; and nodes at level j > 1 receive j + 1 inputs, of which j inputs are the outputs of the previous j nodes in the same skip pathway and the last input is the up-sampled output from the lower skip pathway. The reason that all prior feature maps accumulate and arrive at the current node is because we make use of a dense convolution block along each skip pathway. Fig. 1b further clarifies Eq. 1 by showing how the feature maps travel through the top skip pathway of UNet++.
函数H(·)是一个卷积操作,且紧跟着一个激活函数.U(·)表示一个上采样层,[ ]表示连接层.基本上,级别j=0的节点从编码的先前的层只接收一个输入;级别j=1的节点接收两个输入,它们都来自编码子网络且是两个连续的层;层j>1的节点接收j+1输入,输入j是先前节点j的输出在相同的跳跃路径,而最后一个输入是从更低的跳跃路径的上采样的输出.所有先前的特征图积累并达到正确节点是因为我们沿着每一个跳跃路径上利用一个稠密卷积块.Fig.1b 通过展示特征图如何经历UNet++的最顶端跳跃路径,更加地理清楚Eq.1 .
3.2 Deep supervision 深度监督
We propose to use deep supervision [6] in UNet++, enabling the model to operate in two modes: 1) accurate mode wherein the outputs from all segmentation branches are averaged; 2) fast mode wherein the final segmentation map
is selected from only one of the segmentation branches, the choice of which determines the extent of model pruning and speed gain. Fig. 1c shows how the choice of segmentation branch in fast mode results in architectures of varying
complexity.我们建议在UNet++中使用深度监督[6],使模型能够在两种模式下运行: 1)精确模式,其中输出从所有分割分支平均;2)快速模型,其中最终的分割图只选择分割分支的一个,这个选择确定模型修剪的程度和速度增益.图1c 显示了快速模式下分割分支的选择是如何导致不同复杂度的架构的。
Owing to the nested skip pathways, UNet++ generates full resolution feature maps at multiple semantic levels,
,which are amenable to deep supervision. We have added a combination of binary cross-entropy and dice coefficient as the loss function to each of the above four semantic levels, which is described as:
由于嵌套的跳过路径,UNet++在多个语义级别上形成全分辨率特征图,
是值得深入监督的.我们将二元交叉熵和Dice系数的组合作为损失函数添加到上述四个语义层次中的每一个,描述如下:
where Y^b and Yb denote the flatten predicted probabilities and the flatten ground truths of bth image respectively, and N indicates the batch size
其中Y^b和Yb分别表示bth图像的平坦化预测概率和平坦化基本事实,N表示批大小
In summary, as depicted in Fig. 1a, UNet++ differs from the original U-Net in three ways: 1) having convolution layers on skip pathways (shown in green),which bridges the semantic gap between encoder and decoder feature maps; 2)
having dense skip connections on skip pathways (shown in blue), which improves gradient flow; and 3) having deep supervision (shown in red), which as will be shown in Section 4 enables model pruning and improves or in the worst case achieves comparable performance to using only one loss layer总结,正如图1a描述的UNet++不同于原来U-Net有三个方面:1)在跳跃路径上有卷积层(绿色表示),连接编码器和解码器特征图之间的语义鸿沟;2)在跳跃路径上有密集的跳跃连接(如蓝色所示),这改善了梯度流;3)进行深度监控(如图所示),如第4节所示,可以进行模型修剪和改进,或者在最坏的情况下,可以获得与只使用一个损失层相当的性能.
4 Experiments 实验
Datasets: As shown in Table 1, we use four medical imaging datasets for model evaluation, covering lesions/organs from different medical imaging modalities. For further details about datasets and the corresponding data pre-processing, we refer the readers to the supplementary material.
数据集:表1现实,我们使用了四种医学图像数据集用来模型的评估,包括来自不同医学成像模态的病变/器官.有关数据集和相应数据预处理的进一步详细信息,请参阅补充资料.
Baseline models: For comparison, we used the original U-Net and a customized wide U-Net architecture. We chose U-Net because it is a common performance baseline for image segmentation. We also designed a wide U-Net with similar number of parameters as our suggested architecture. This was to ensure that the performance gain yielded by our architecture is not simply due to increased number of parameters. Table 2 details the U-Net and wide U-Net architecture
基准模型:为了比较,我们使用了原始的U-Net和wide U-Net.我们选择U-Net,因为它是图像分割的通用性能基准.我们也设计一个具有相似参数的数量wide U-Net作为我们的建议的架构.这是为了确保我们的体系结构产生的性能收益不仅仅是由于参数数量的增加。表2详细说明了U-Net和wide U-Net体系结构
Implementation details: We monitored the Dice coefficient and Intersectionover Union (IoU), and used early-stop mechanism on the validation set. We also used Adam optimizer with a learning rate of 3e-4. Architecture details for UNet and wide U-Net are shown in Table 2. UNet++ is constructed from the original U-Net architecture. All convolutional layers along a skip pathway (Xi;j) use k kernels of size 3×3 (or 3×3×3 for 3D lung nodule segmentation) where k = 32 × 2i. To enable deep supervision, a 1×1 convolutional layer followed by a sigmoid activation function was appended to each of the target nodes: fx0;j j j 2 f1; 2; 3; 4gg. As a result, UNet++ generates four segmentation maps given an input image, which will be further averaged to generate the final segmentation map. More details can be founded at github.com/Nested-UNet.
实现细节:我们监控了Dice系数和Intersectionover Union (IoU),并在验证集上使用了提前终止机制。我们还使用了学习率为3e-4的Adam optimizer。UNet和wide U-Net的架构细节如表2所示。UNet++是由原来的U-Net架构构建的。所有沿skip pathway (Xi;j)的convolutional layers使用大小为3×3(或3×3×3用于三维肺结节分割)的k个kernel,其中k = 32×2i。为了实现深度监控,在每个目标节点上附加1×1的卷积层和一个sigmoid激活函数:fx0;j j j 2 f1;2;3;4 gg。结果,UNet++生成四个分割地图给定一个输入图像,这将进一步平均,以生成最终的分割地图。更多详情请访问github.com/Nested-UNet。
Fig. 2: Qualitative comparison between U-Net, wide U-Net, and UNet++, showing segmentation results for polyp, liver, and cell nuclei datasets (2D-only for a distinct visualization).
图2:U-Net、wide U-Net和UNet++的定性比较,显示了息肉、肝脏和细胞核数据集(2D-only for a)的分割结果不同的可视化)。
Results: Table 3 compares U-Net, wide U-Net, and UNet++ in terms of the number parameters and segmentation accuracy for the tasks of lung nodule segmentation, colon polyp segmentation, liver segmentation, and cell nuclei segmentation. As seen, wide U-Net consistently outperforms U-Net except for liver segmentation where the two architectures perform comparably. This improvement is attributed to the larger number of parameters in wide U-Net. UNet++ without deep supervision achieves a significant performance gain over both UNet and wide U-Net, yielding average improvement of 2.8 and 3.3 points in IoU. UNet++ with deep supervision exhibits average improvement of 0.6 points over UNet++ without deep supervision. Specifically, the use of deep supervision leads to marked improvement for liver and lung nodule segmentation, but such improvement vanishes for cell nuclei and colon polyp segmentation. This is because polyps and liver appear at varying scales in video frames and CT slices; and thus, a multi-scale approach using all segmentation branches (deep supervision) is essential for accurate segmentation. Fig. 2 shows a qualitative comparison between the results of U-Net, wide U-Net, and UNet++.
结果:表3比较了U-Net、wide U-Net和UNet++在肺结节、结肠息肉、肝脏和细胞核的数量参数和分割准确性。从图中可以看出,wide U-Net的性能一直都比U-Net好,除了肝脏部分,这两种架构的性能比较接近。这种改进是由于wide U-Net中参数数目较多。UNet++在没有深度监督的情况下,在UNet和宽U-Net上都获得了显著的性能提升,IoU平均提高2.8和3.3分。有深度监督的UNet++平均比没有深度监督的UNet++提高0.6分。具体来说,使用深度监督可以明显改善肝、肺结节的分割,但这种改善消失在细胞核和结肠息肉的分割上。这是因为息肉和肝脏在视频帧和CT切片中出现的范围不同;因此,一个多尺度的方法使用所有的分割分支(深度监督)是必要的准确分割。图2是U-Net、wide U-Net和UNet++的定性比较结果。
Fig. 3: Complexity, speed, and accuracy of UNet++ after pruning on (a) cell nuclei, (b) colon polyp, (c) liver, and (d) lung nodule segmentation tasks respectively. The inference time is the time taken to process 10k test images using one NVIDIA TITAN X (Pascal) with 12 GB memory.
图3:分别对(a)细胞核、(b)结肠息肉、(c)肝脏和(d)肺结节进行剪枝后的UNet++的复杂度、速度和准确性。预测时间是使用一个NVIDIA TITAN X (Pascal)和12gb内存处理10k测试图像所花费的时间。
Model pruning: Fig. 3 shows segmentation performance of UNet++ after applying different levels of pruning. We use UNet++ Li to denote UNet++ pruned at level i (see Fig. 1c for further details). As seen, UNet++ L3 achieves on average 32.2% reduction in inference time while degrading IoU by only 0.6 points. More aggressive pruning further reduces the inference time but at the cost of significant accuracy degradation
模型剪枝:UNet++经过不同程度的剪枝后,其分割性能如图3所示。我们用UNet++ Li表示第i级修剪的UNet++(详见图1c)。由此可见,UNet++ L3平均推理时间减少32.2%,IoU仅减少0.6分。更积极的剪枝进一步减少推断时间,但代价是显著的准确性下降
5 Conclusion 结论
To address the need for more accurate medical image segmentation, we proposed UNet++. The suggested architecture takes advantage of re-designed skip pathways and deep supervision. The re-designed skip pathways aim at reducing the semantic gap between the feature maps of the encoder and decoder subnetworks, resulting in a possibly simpler optimization problem for the optimizer to solve. Deep supervision also enables more accurate segmentation particularly for lesions that appear at multiple scales such as polyps in colonoscopy videos. We evaluated UNet++ using four medical imaging datasets covering lung nodule segmentation, colon polyp segmentation, cell nuclei segmentation, and liver segmentation. Our experiments demonstrated that UNet++ with deep supervision achieved an average IoU gain of 3.9 and 3.4 points over U-Net and wide U-Net, respectively
为了解决更精确的医学图像分割的需要,我们提出了UNet++。建议的架构利用了重新设计的跳跃路径和深度监督。重新设计的跳跃路径旨在减少编码器和解码器子网的特征映射之间的语义差距,从而为优化器解决一个可能更简单的优化问题。深度监控还可以更准确地分割病变,特别是出现在多个尺度上的病变,如结肠镜检查视频中的息肉。我们使用四组医学成像数据集对UNet++进行了评估,包括肺结节分割、结肠息肉分割、细胞核分割和肝脏分割。我们的实验表明,UNet++在深度监管下,比U-Net和宽U-Net的平均IoU增益分别为3.9和3.4分
Acknowledgments This research has been supported partially by NIH under Award Number R01HL128785, by ASU and Mayo Clinic through a Seed Grant and an Innovation Grant. The content is solely the responsibility of the authors and does not necessarily represent the official views of NIH
本研究得到美国国立卫生研究院(NIH) R01HL128785号资助,美国亚利桑那州立大学(ASU)和梅奥诊所(Mayo Clinic)通过种子基金和创新基金的部分支持。内容完全是作者的责任,不一定代表NIH的官方观点
References
![[论文翻译]UNet++: A Nested U-Net Architecture for Medical Image Segmentation_第1张图片](http://img.e-com-net.com/image/info8/6f39003da5fc46dab75ca45728c63413.jpg)

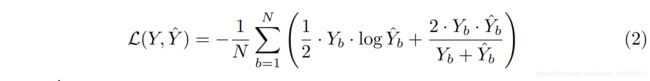
![[论文翻译]UNet++: A Nested U-Net Architecture for Medical Image Segmentation_第2张图片](http://img.e-com-net.com/image/info8/075e00ba1a824faa94cc6f61709ab998.jpg)
![[论文翻译]UNet++: A Nested U-Net Architecture for Medical Image Segmentation_第3张图片](http://img.e-com-net.com/image/info8/f805c1db2ff9412e8dbd2ec18c594906.jpg)
![[论文翻译]UNet++: A Nested U-Net Architecture for Medical Image Segmentation_第4张图片](http://img.e-com-net.com/image/info8/077ff595f6aa43b0977946e4de80098e.jpg)
![[论文翻译]UNet++: A Nested U-Net Architecture for Medical Image Segmentation_第5张图片](http://img.e-com-net.com/image/info8/b83e1b6b8ab34090a1f9c569a3ca7793.jpg)