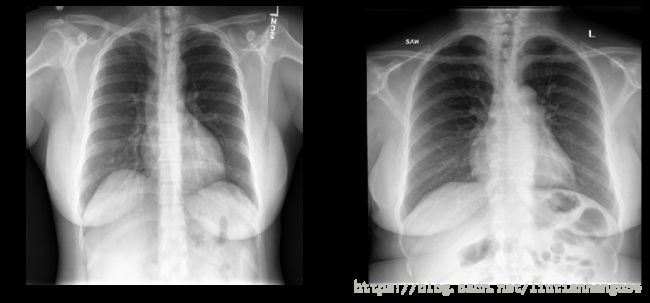
kaggle 入门系列翻译(四) RSNA 肺炎预测
https://github.com/mdai/ml-lessons/blob/master/lesson3-rsna-pneumonia-detection-kaggle.ipynb
上述是官方提供的一个教学,点进去之后共有四个章节,本文先翻译第一个章节:
针对使用深度学习进行医疗图像识别
第一课:胸部和腹部x光的分类
这是对用于医学图像分类的实用机器学习的高级介绍。本教程的目标是建立一个深度学习分类器来精确区分胸部和腹部x光。该模型使用从Open-i中获得的75幅图像去识别图像进行训练。
使用MD.ai注释器查看DICOM图像,并创建图像级别注释。然后使用MD.ai python客户端库下载图像和注释,准备数据集,然后用于训练模型进行分类。
- Original public github repo is based on: https://github.com/ImagingInformatics/machine-learning
- MD.ai python client libray URL: https://github.com/mdai/mdai-client-py
- MD.ai documentation URL: https://docs.md.ai/
课程目录如下:
-
Lesson 1. Classification of chest vs. adominal X-rays using TensorFlow/Keras Github Annotator
-
Lesson 2. Lung X-Rays Semantic Segmentation using UNets. Github Annotator
-
Lesson 3. RSNA Pneumonia detection using Kaggle data format Github Annotator
-
Lesson 3. RSNA Pneumonia detection using MD.ai python client library Github Annotator
首先安装 mdai 模块:
pip install mdai创建一个mdai客户端
mdai客户机需要一个访问令牌,它将您验证为用户。要创建新的令牌或选择现有令牌,请在指定的MD.ai域中导航到用户设置页面上的“Personal Access token”选项卡(例如,public.md.ai)
mdai_client = mdai.Client(domain='public.md.ai', access_token="")建立项目
通过传递项目id来定义您可以访问的项目。项目id可以在URL中找到,格式如下:https://public.md.ai/annotator/project/{project_id}。
例如,project_id为PVq9raBJ (https://public.md.ai/annotator/project/PVq9raBJ)。
指定可选路径作为数据目录(如果留空,将默认为当前工作目录)。
p = mdai_client.project('PVq9raBJ', path='./lesson1-data')设置id
为了准备数据集,选定的标签id必须由项目#set_label_ids方法显式地设置。
p.show_label_groups()
# this maps label ids to class ids as a dict obj
labels_dict = {'L_38Y7Jl':0, # Abdomen
'L_z8xEkB':1, # Chest
}
print(labels_dict)
p.set_labels_dict(labels_dict)Label Group, Id: G_3lv, Name: Default group Labels: Id: L_38Y7Jl, Name: Abdomen Id: L_z8xEkB, Name: Chest
{'L_38Y7Jl': 0, 'L_z8xEkB': 1}
创建训练集和测试集
p.show_datasets()
# create training dataset
train_dataset = p.get_dataset_by_name('TRAIN')
train_dataset.prepare()
train_image_ids = train_dataset.get_image_ids()
print(len(train_image_ids))
# create the validation dataset
val_dataset = p.get_dataset_by_name('VAL')
val_dataset.prepare()
val_image_ids = val_dataset.get_image_ids()
print(len(val_image_ids))Datasets: Id: D_8ogmzN, Name: TRAIN Id: D_OoJ98E, Name: VAL Id: D_8oAvmQ, Name: TEST
65 10
展示部分图片
# visualize a few train images
mdai.visualize.display_images(train_image_ids[:2], cols=2)
mdai.visualize.display_images(val_image_ids[:2], cols=2)使用keras进行训练和验证
from keras import applications
from keras.models import Model, Sequential
from keras.layers import Dropout, Flatten, Dense, GlobalAveragePooling2D
from keras.optimizers import Adam
from keras.callbacks import EarlyStopping, ModelCheckpoint
# Define model parameters
img_width = 192
img_height = 192
epochs = 20
params = {
'dim': (img_width, img_height),
'batch_size': 5,
'n_classes': 2,
'n_channels': 3,
'shuffle': True,
}
base_model = applications.mobilenet.MobileNet(weights='imagenet', include_top=False, input_shape=(img_width, img_height, 3))
model_top = Sequential()
model_top.add(GlobalAveragePooling2D(input_shape=base_model.output_shape[1:], data_format=None))
model_top.add(Dense(256, activation='relu'))
model_top.add(Dropout(0.5))
model_top.add(Dense(2, activation='softmax'))
model = Model(inputs=base_model.input, outputs=model_top(base_model.output))
model.compile(optimizer=Adam(lr=0.0001, beta_1=0.9, beta_2=0.999, epsilon=1e-08,decay=0.0),
loss='categorical_crossentropy', metrics=['accuracy'])
from mdai.utils import keras_utils
train_generator = keras_utils.DataGenerator(train_dataset, **params)
val_generator = keras_utils.DataGenerator(val_dataset, **params)
import tensorflow as tf
config = tf.ConfigProto()
config.gpu_options.allow_growth = True
# Set callback functions to early stop training and save the best model so far
callbacks = [
EarlyStopping(monitor='val_loss', patience=2, verbose=2),
ModelCheckpoint(filepath='best_model.h5', monitor='val_loss',
save_best_only=True, verbose=2)
]
history = model.fit_generator(
generator=train_generator,
epochs=epochs,
callbacks=callbacks,
verbose=1,
validation_data=val_generator,
use_multiprocessing=True,
workers=6)
import matplotlib.pyplot as plt
print(history.history.keys())
plt.figure()
plt.plot(history.history['acc'], 'orange', label='Training accuracy')
plt.plot(history.history['val_acc'], 'blue', label='Validation accuracy')
plt.plot(history.history['loss'], 'red', label='Training loss')
plt.plot(history.history['val_loss'], 'green', label='Validation loss')
plt.legend()
plt.show()dict_keys(['val_loss', 'val_acc', 'loss', 'acc'])
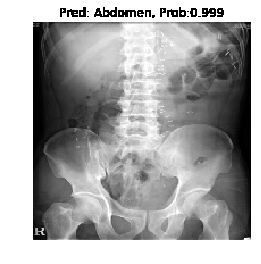
创建测试集
model.load_weights('best_model.h5')
test_dataset = p.get_dataset_by_name('TEST')
test_dataset.prepare()
import numpy as np
#from skimage.transform import resize
from PIL import Image
for image_id in test_dataset.image_ids:
image = mdai.visualize.load_dicom_image(image_id, to_RGB=True)
image = Image.fromarray(image)
image = image.resize((img_width, img_height))
x = np.expand_dims(image, axis=0)
y_prob = model.predict(x)
y_classes = y_prob.argmax(axis=-1)
title = 'Pred: ' + test_dataset.class_id_to_class_text(y_classes[0]) + ', Prob:' + str(round(y_prob[0][y_classes[0]], 3))
plt.figure()
plt.title(title)
plt.imshow(image)
plt.axis('off')
plt.show()