Plant Leaves Classification: A Few-Shot Learning Method Based on Siamese Network
The article directories
- Abstract
- Introduction
- PROPOSED CNN STRUCTURE
- INITIAL CNN ANALYSIS
- EXPERIMENTAL STRUCTURE AND ALGORITHMS
- MATERIALS AND METHODS
- DATASET
- PREPROCESSING OF IMAGES
- FEW-SHOT LEARNING AND DISTANCE TRAINING STRATEGY
- CONVOLUTIONAL NEURAL NETWORKS
- NEAREST NEIGHBOR CLASSIFICATION
- OVERALL EXPERIMENTAL PROCEDURE
- EXPERIMENTS AND RESULTS
- EXPERIMENTAL DATASET AND SSO VERIFICATION
- COMPARISON WITH OTHER CNN FRAMEWORKS
- COMPARISON WITH SEMISUPERVISED METHODS
- CONCLUSION AND FUTURE WORK
- REFERENCES
Abstract
In this paper, a few-shot learning method based on the Siamese network framework isproposed to solve a leaf classification problem with a small sample size.
- First, the features of two differentimages are extracted by a parallel two-way convolutional neural network with weight sharing.
- Then, the network uses a loss function to learn the metric space, in which similar leaf samples are close to each other anddifferent leaf samples are far away from each other.
- In addition, a spatial structure optimizer (SSO) methodfor constructing the metric space is proposed, which will help to improve the accuracy of leaf classification.
- Finally, a k-nearest neighbor (kNN) classifier is used to classify leaves in the learned metric space.
Datasets:
The open access Flavia, Swedish andLeafsnap datasets are used to evaluate the performance of the method
The experimental results:
the proposed method can achieve a high classification accuracy with a small size of supervised samples.
INDEX TERMS:
Leaf classification, few-shot learning, convolutional neural network, Siamese network.
Introduction
In past studies, features for plant identification were usu-ally selected from plant organs such as leaves, flowers, fruits,and stems, among which the leaves of plants are the most representative and easiest to obtain.[with a semisupervised method [5] in machine learning]
Recently, due to the excellent performance of deep learning convolutional neural networks in the fieldof computer vision, they has become the main means tosolve the problems of image classification, image recogni-tion and semantic segmentation [14]–[16].
the disadvantages of deep learning:
- The premise of high classification accuracy is that thenetwork has sufficient supervised learning samples, which isusually very difficult.
Therefore, theconcept of few-shot learning is proposed.
Therefore we constructed a structure based ona Siamese network [21]–[23] to extract the characteristics ofplant leaves and classify them.
The main contributions:
- A method based on the Siamese network structure is proposed to construct a metric space for leaf classifica-tion
- A spatial structure optimizer is proposed to improve the speed and performance of measuring the spatia lformation process.
- Experimental shows that this method can effectively classify leaves with a smallnumber of supervised samples.
PROPOSED CNN STRUCTURE
INITIAL CNN ANALYSIS
The magnitude of the model and the number of calculations were measured with FLOPs and params, respectively.
According to the experimental results, this paper proposes using the inception-v4 structure.Although a deep DenseNet has better performance in modelcomplexity, DenseNet consumes much memory, which is not conducive to the implementation of the project.

EXPERIMENTAL STRUCTURE AND ALGORITHMS
The training structure for each batch is shown in Fig. 1.
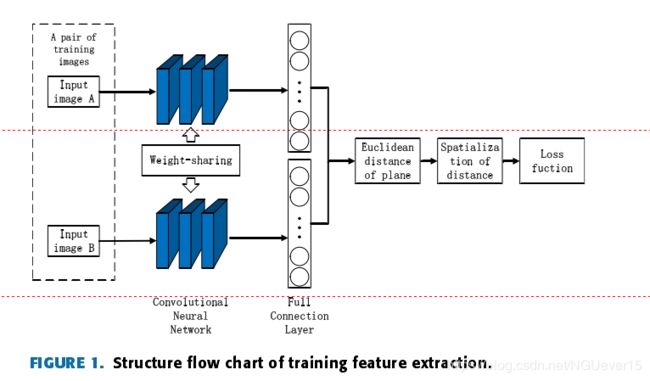
In the design of feature extractors, we refer to andimprove the Inception structure to increase the adaptability of the network to the input image scale, and reduce the phenomena of gradient disappearance and overfitting, thus the adverse impact caused by the sample itself
what is Inception structure ?
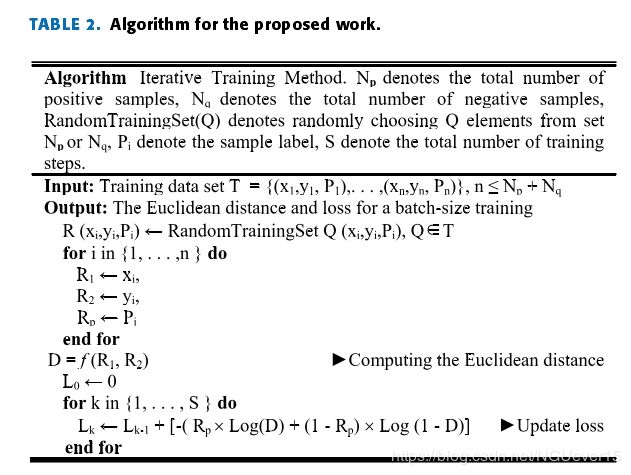
The last part of the structure uses the logistic regression lossfunction to measure the similarity between input image pairs.Table 2 shows the proposed steps of the algorithm.
MATERIALS AND METHODS
DATASET
we choose the Flavia, Swedishand Leafsnap datasets for the training and test sets.
the numbers of training images for each supervised sample are different(5-20, and the increment is 5)
All other images that are notselected as monitoring samples will constitute a verification set to evaluate the algorithm.
It is worth noting that a single training sample consists of two pictures.
If they belong to the same category, it is called a positive sample, and the sampleis labeled with a 1
| type | positive sample | negative sample |
|---|---|---|
| lable | 1 | 0 |
Fig. 2 shows some samples of the Flavia, Swedish andLeafsnap datasets.
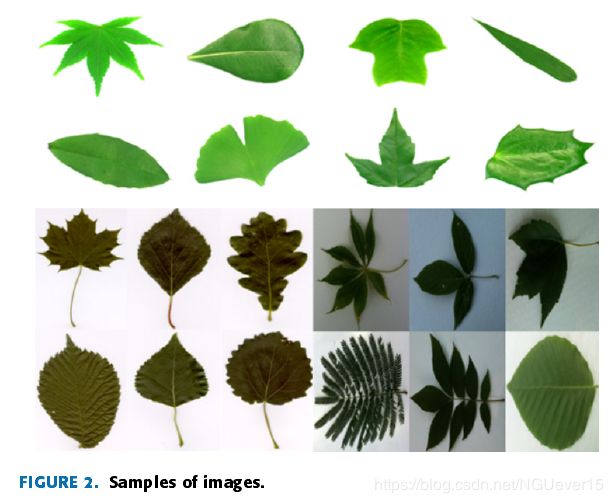
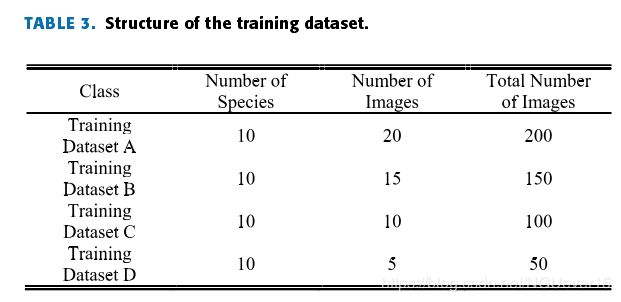
Table 3 shows the composition of the four subsets required for each dataset experiment
PREPROCESSING OF IMAGES
image preprocessing:
In this study, the sizes of all the images inthe dataset were uniformly adjusted to 112×112 pixels bycentral clipping, which was automatically completed by thecomputer through the OpenCV framework and Python script.
Eq. (1) gives the center square clipping method for imagescaling.
def SquareResize(Img, New_height, New_width) (1)
FEW-SHOT LEARNING AND DISTANCE TRAINING STRATEGY
The Siamese network structurecan map the similarity relationship between different imagesinto a metric space so that the samples belonging to the samecategory can be as close as possible, and the samples belong-ing to different categories can be as far away as possible.
The method used in this paper is trained in a supervised way, and the samples are extracted by a two-way convolution neuralnetwork.
Then, the Euclidean distance between features is calculated by a metric-based method: the closer the distanceis, the more similar the samples are.
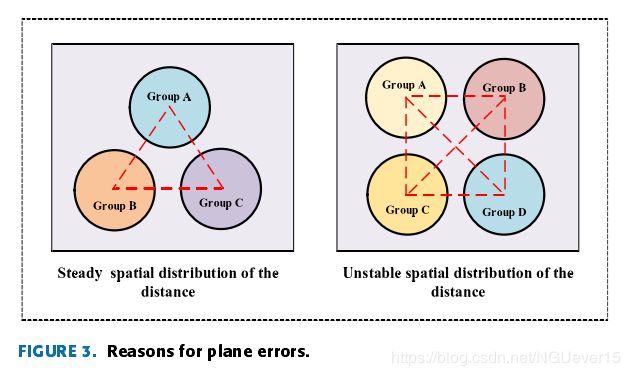
Reasons for plane errors:
three similar samples will be in the metric space mappingplane and form a stable distribution of an equilateral triangle, but the four similar samples will endanger the stability ofthe original mapping plane, causing its distribution to be asquare, which can force the distances between samples in the diagonal positions to increase, as shown in Fig. 3 (the red dotted line represents the average distance between the sample classes).
Therefore, we propose the SSO that acts on the process ofmetric space formation.
The spatial distribution of the dis-tance is achieved by using the stability of the spatial structureof a regular tetrahedron to accelerate the training convergence speed and improve the accuracy, as shown in Fig. 4.

When sample D meets the SSO condition, the distance of sample D will be mapped into the β plane. In the subsequent training, only the distances betweensamples A, B and C related to it will be trained.
In the k-nearest neighbor classifier, the distance between sample Dand other unassociated samples E is replaced by formula eq. (2).
distance(DE) = mean(distance(AE) + distance(BE)+distance(CE)) (2)
when multiple SSO conditions aretriggered, the distance is not calculated between all samples distributed in the beta plane during training
The trigger condition of the SSO structure is calculated byeq. (3).
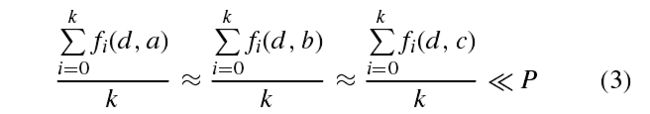
where k denotes the minimum number of sample distances calculations to be met, f (d, a), f (d, b), and f (d, c) are theEuclidean distance functions between samples, and P is theEuclidean distance value that satisfies the trigger condition
to distinguishsamples in different planes, it is necessary to mark the map-ping plane of the samples in the training process, which is expressed as eq. (4) in the program.
fy = (Distance, n) (4)
where Distance is the Euclidean distance function of the samples, and n is the distribution plane.
The loss function is shown in eq. (5).
![]()
where L is the loss function and f is the label of the inputpair., if the input images are from the same class, f = 1,;otherwise, f = 0,; fy is the European distance for the trainingpair.
CONVOLUTIONAL NEURAL NETWORKS
Generally, this architecture is composed of five parts: theinput layer, convolution layer, pooling layer, full connectionlayer, and output layer.
Fig. 5 shows our pro-posed CNN model for extracting and processing leaf features.This model is inspired by the structure of GoogLeNet.

GoogLeNet can allocate computing resources better and extract more features than other models under the samecomputation amount
GoogLeNet can solve the gradient disappearance, gradient explosion and other problemscaused by the ultradeep network.
GoogLeNet introducedthe concept of the ‘‘Inception module’’, the idea of which is to use relatively dense components to approximate the optimal local sparse structure.
NEAREST NEIGHBOR CLASSIFICATION
As shown in Fig. 7, during the test phase, the tested sampleand the supervised sample are extracted by the convolutionalneural network.
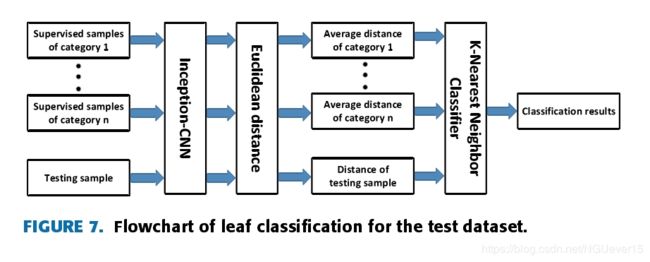
through a simple kNN classifier, the classification task canbe completed by comparing and analyzing the Euclideandistance between the samples to be tested and different kinds of supervised samples, as shown in Fig. 8.

OVERALL EXPERIMENTAL PROCEDURE
The overall experimental procedure is shown in Fig. 6.
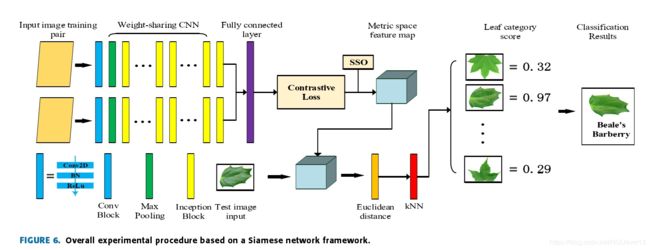
the contrast loss function is used to form the metric space with the SSO.
When theimage is input, the network calculates the Euclidean distancebetween the sample to be tested and the known species in the metric space, and outputs the similarity score through the kNN classifier.
EXPERIMENTS AND RESULTS
Devices:
All the experiments were conducted on a laptop with an IntelCore i7-6700HQ processor (2.6 GHz) and an Nvidia GeforceGTX 1060 6 GB graphics card. The laptop has 16 GB ofmemory.
The training and testing work was implementedusing the open-source software framework TensorFlow.
Therecommended parameters for the CNN were set as follows:
the learning rate was set to 0.001, the dropout rate was set to 0.5, the training step length was set to 30000, and the batchsize was set to 8.
EXPERIMENTAL DATASET AND SSO VERIFICATION
It should be noted that thenumber of negative training samples is larger than that of thepositive ones, so we need to randomly remove some negativetraining samples.
Table 4 shows the number of positive sam-ples and negative samples in the four subsets of the Flavia,Swedish and Leafsnap datasets.
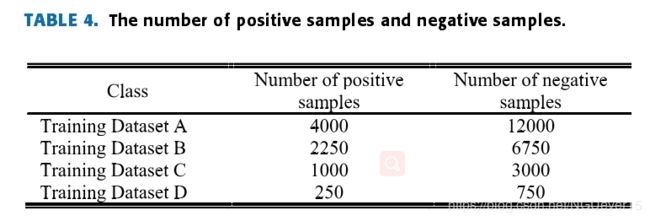
Fig. 9 shows four leaves that meet the requirementsof a particular training set: in some ways, it is difficult fornonprofessionals to classify these leaves accurately.

Fig. 10 shows the variation in the loss curve with the SSO and without the SSO.
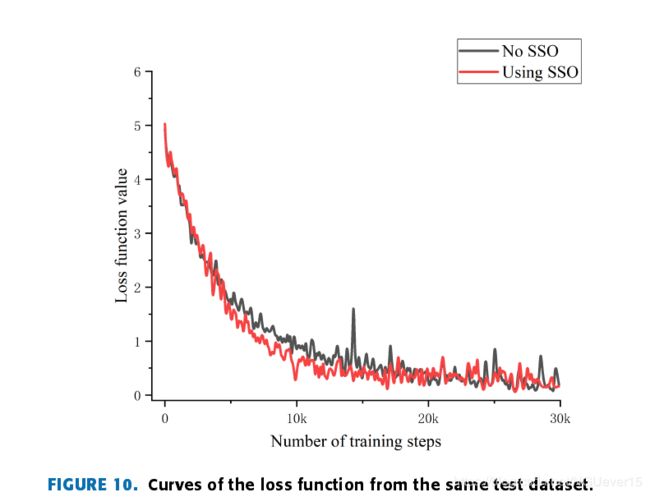
As shown in the figure:
- the SSO loss curveconverges faster and the descent process is smoother beforethe 20000 steps.
- In addition, at the end of training, the stability of the SSO loss curve is better.
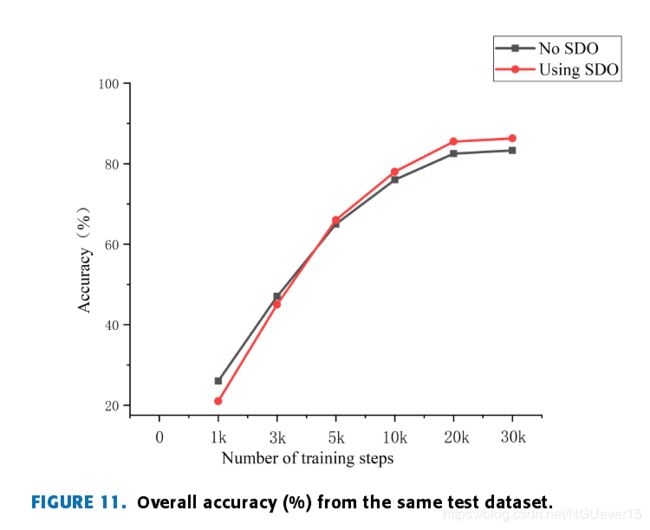
Fig. 11 shows the classification accuracy results of the kNN classifier on the same test datasetwith and without the SSO.
According to the curve analy-sis, as the number of training steps increases, the networkadvantages of SSO training gradually emerge, and a highclassification accuracy is maintained in the later stages.
Fig. 12 is a metric space without the SSO.
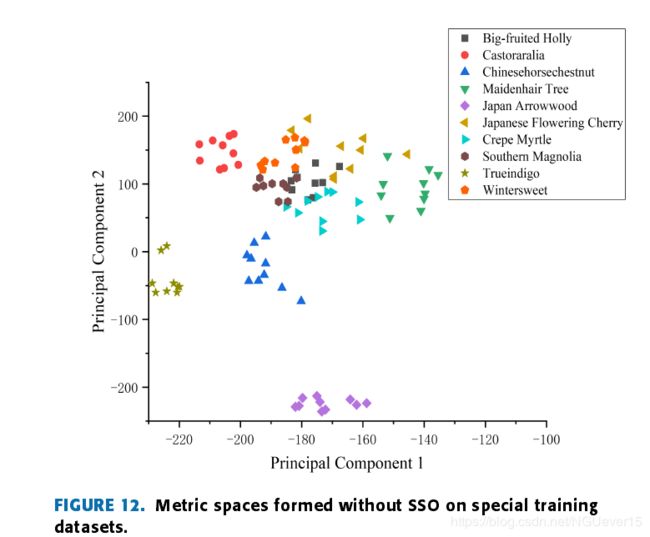
Fig. 13 is a metric space formed using the SSO.
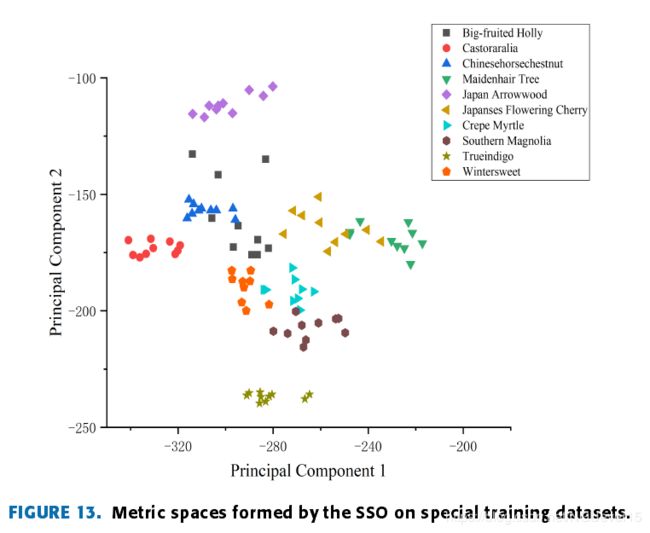
it is wellseparated from the similar Japanese flowering cherry, crepemyrtle and wintersweet samples.
the distribution of the metric space tends to be morereasonable, the distributions of the same kind of leaves ismore concentrated, and the fault-tolerance rate is higher in theprocess of kNN classification, so the classification accuracyis higher.
COMPARISON WITH OTHER CNN FRAMEWORKS
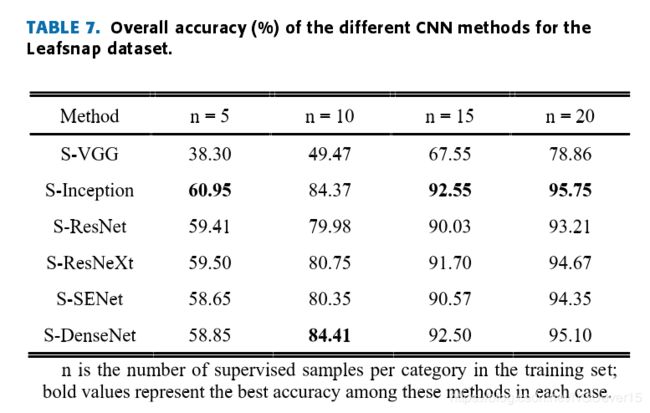
Table 5,Table 6 and Table 7 lists the test results. Compared withother CNN frameworks, the adjusted Siamese + Incep-tion (S-Inception) network can provide competitive results,and the results show that the S-Inception combination can achieve good accuracy when the training sample settingsare appropriate.
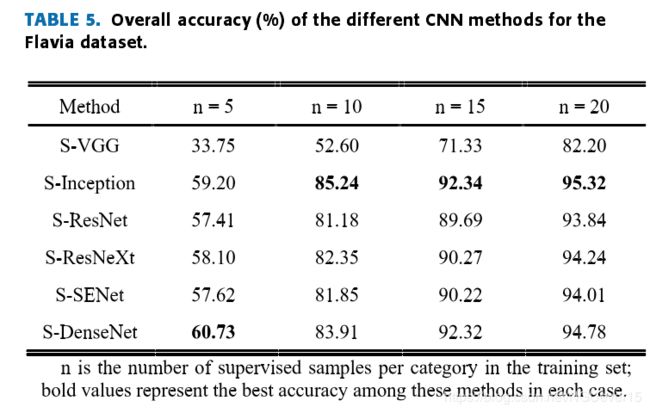
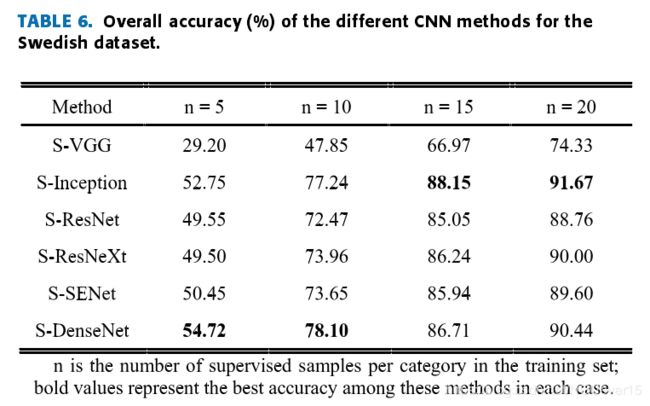
When the number of samples isinsufficient, the deep network gradient disappears seriously,and serious overfitting will occur.
Overall,the test results show that it is good to combine the Siamesenetwork structure with different CNN frameworks to realizethe few-shot learning classification method of leaves, whichfurther proves the effectiveness of the method. The accuracyis computed by eq. (6).
COMPARISON WITH SEMISUPERVISED METHODS
Table 8, Table 9 and Table 10 showthe results of the comparative tests. As we can see fromthe table, all the methods improve the accuracy when thenumber of supervised samples increases, but the S-Inceptionmethod improves faster, because the generalization abilityof the S-Inception structure is better, and the width of thestructure makes it possible to extract more features when thenumber of samples increases.


The experimental results show that this methodperforms better than some semisupervised methods.
CONCLUSION AND FUTURE WORK
In this paper, an improved convolutional neural networkstructure is proposed to solve the problem of leaf classifica-tion in the case of small samples
The key to this method is toextract image features by using a convolutional neural net-work and construct the metric space by using the concept ofsimilarity between different image features.
The experimental results also show that when the number of training samplesis 20, the classification accuracy of this method is the high-est, 95.32%, 91.37% and 91.75% accuracies are obtainedfrom the Flavia, Swedish and Leafsnap datasets, respectively.These results are competitive in the deep learning classifica-tion field.
further research is needed toimprove the generalization ability of the model. In the future,our method will be applied to more datasets and few-shotclassification tasks.
REFERENCES
[1] S. L. Pimm, C. N. Jenkins, R. Abell, T. M. Brooks, J. L. Gittleman,L. N. Joppa, P. H. Raven, C. M. Roberts, and J. O. Sexton, ‘‘The biodiver-sity of species and their rates of extinction, distribution, and protection,’’Science, vol. 344, no. 6187, May 2014, Art. no. 7246752. doi: 10.1126/sci-ence.1246752.
[2] J. Wäldchen, M. Rzanny, M. Seeland, and P. Mäder, ‘‘Automated plantspecies identification—Trends and future directions,’’ PLoS Comput.Biol., vol. 14, no. 4, Apr. 2018, Art. no. e1005993. doi: 10.1371/jour-nal.pcbi.1005993.
[3] B. Dayrat, ‘‘Towards integrative taxonomy,’’ Biol. J. Linnean Soc., vol. 85,no. 85, pp. 407–415, Jul. 2005. doi: 10.1111/j.1095-8312.2005.00503.x.
[4] Y. Sun, Y. Liu, G. Wang, and H. Zhang, ‘‘Deep learning for plant iden-tification in natural environment,’’ Comput. Intell. Neurosci., vol. 2017,May 2017, Art. no. 7361042. doi: 10.1155/2017/7361042
[5] V. Narayan and G. Subbarayan, ‘‘An optimal feature subset selection usingGA for leaf classification,’’ Int. Arab J. Inf. Technol., vol. 11, no. 5,pp. 447–451, Sep. 2014. doi: 10.1109/TIT.2014.2344251.
[6] H. Qi, T. Shuo, and S. Jin, ‘‘Leaf characteristics-based computer-aidedplant identification model,’’ J. Zhejiang Forestry College, vol. 20, no. 3,pp. 281–284, 2003. doi: 10.1023/A:1022289509702.
[7] C. Zhao, S. S. F. Chan, W.-K. Cham, and L. M. Chu, ‘‘Plant identifi-cation using leaf shapes—A pattern counting approach,’’ Pattern Recog-nit., vol. 48, no. 10, pp. 3203–3215, Oct. 2015. doi: 10.1016/j.patcog.2015.04.004.
[8] A. Aakif and M. F. Khan, ‘‘Automatic classification of plants basedon their leaves,’’ Biosyst. Eng., vol. 139, pp. 66–75, Nov. 2015.doi: 10.1016/j.biosystemseng.2015.08.003.
[9] J. R. Kala and S. Viriri, ‘‘Plant specie classification using sinuosity coeffi-cients of leaves,’’ Image Anal. Stereol., vol. 37, no. 2, pp. 119–126, 2018.doi: 10.5566/ias.1821.
[10] H. Kolivand, B. M. Fern, T. Saba, M. S. M. Rahim, and A. Rehman, ‘‘A newleaf venation detection technique for plant species classification,’’ ArabianJ. Sci. Eng., vol. 44, no. 4, pp. 3315–3327, Apr. 2019. doi: 10.1007/s13369-018-3504-8.
[11] S. Zhang, C. Zhang, Z. Wang, and W. Kong, ‘‘Combining sparse repre-sentation and singular value decomposition for plant recognition,’’ Appl.Soft Comput., vol. 67, pp. 164–171, Jun. 2018. doi: 10.1016/j.asoc.2018.02.052.
[12] M. B. H. Rhouma, J. Žunić, and M. C. Younis, ‘‘Moment invariants formulti-component shapes with applications to leaf classification,’’ Com-put. Electron. Agricult., vol. 142, pp. 326–337, Nov. 2017. doi: 10.1016/j.compag.2017.08.029.
[13] S. J. Kho, S. Manickam, S. Malek, M. Mosleh, and S. K. Dhillon,‘‘Automated plant identification using artificial neural network and supportvector machine,’’ Frontiers Life Sci., vol. 10, no. 1, pp. 98–107, 2017.doi: 10.1080/21553769.2017.1412361
.[14] Y. LeCun, Y. Bengio, and G. Hinton, ‘‘Deep learning,’’ Nature, vol. 521,no. 7553, pp. 436–444, May 2015. doi: 10.1038/nature14539.
[15] H.-C. Shin, H. R. Roth, M. Gao, L. Lu, Z. Xu, I. Nogues, J. Yao,D. Mollura, and R. M. Summers, ‘‘Deep convolutional neural networks forcomputer-aided detection: CNN architectures, dataset characteristics andtransfer learning,’’ IEEE Trans. Med. Imag., vol. 35, no. 5, pp. 1285–1298,May 2016. doi: 10.1109/TMI.2016.2528162
[16] U. P. Singh, S. S. Chouhan, S. Jain, and S. Jain, ‘‘Multilayer convo-lution neural network for the classification of mango leaves infectedby anthracnose disease,’’ IEEE Access, vol. 7, pp. 43721–43729, 2019.doi: 10.1109/ACCESS.2019.2907383.
[17] J. Hu, Z. Chen, M. Yang, R. Zhang, and Y. Cui, ‘‘A multiscale fusionconvolutional neural network for plant leaf recognition,’’ IEEE SignalProcess. Lett., vol. 25, no. 6, pp. 853–857, Jun. 2018. doi: 10.1109/LSP.2018.2809688.
[18] S. A. Pearline, V. S. Kumar, and S. Harini, ‘‘A study on plant recognitionusing conventional image processing and deep learning approaches,’’J. Intell. Fuzzy Syst., vol. 36, no. 3, pp. 1997–2004, 2019.doi: 10.3233/JIFS-169911.
[19] T. K. N. Thanh, Q. B. Truong, Q. D. Truong, and H. H. Xuan, ‘‘Depthlearning with convolutional neural network for leaves classifier basedon shape of leaf vein,’’ in Proc. Asian Conf. Intell. Inf. Database Syst.(ACIIDS), 2018, pp. 575–585. doi: 10.1007/978-3-319-75417-8_53.
[20] J. Snell, K. Swersky, and R. Zemel, ‘‘Prototypical networks for few-shotlearning,’’ in Proc. Adv. Neural Inf. Process. Syst., 2017, pp. 4077–4087.[Online]. Available: https://arxiv.org/abs/1703.05175.
[21] Z.-Y. Gao, H.-X. Xie, J.-F. Li, and S.-L. Liu, ‘‘Spatial-structuresiamese network for plant identification,’’ Int. J. Pattern Recognit. Artif.Intell., vol. 32, no. 11, Nov. 2018, Art. no. 1850035. doi: 10.1142/S0218001418500350.
[22] S. Chopra, R. Hadsell, and Y. LeCun, ‘‘Learning a similarity metricdiscriminatively, with application to face verification,’’ in Proc. IEEEComput. Soc. Conf. Comput. Vis. Pattern Recognit. (CVPR), Jun. 2005,pp. 539–546. doi: 10.1109/CVPR.2005.202.
[23] J. Bromley, J. W. Bentz, L. Bottou, I. Guyon, Y. Lecun, C. Moore,E. Säckinger, and R. Shah, ‘‘Signature verification using a ‘Siamese’ timedelay neural netwoRK,’’ Int. J. Pattern Recognit. Artif. Intell., vol. 7, no. 4,pp. 669–688, 1993. doi: 10.1142/S0218001493000339.
[24] S. G. Wu, F. S. Bao, E. Y. Xu, Y.-X. Wang, Y.-F. Chang and Q.-L. Xiang,‘‘A leaf recognition algorithm for plant classification using probabilisticneural network,’’ in Proc. IEEE Int. Symp. Signal Process. Inf. Technol.,Dec. 2007, pp. 11–16. doi: 10.1109/ISSPIT.2007.4458016.
[25] O. Soderkvist, ‘‘Computer vision classification of leaves from Swedishtrees,’’ Teknik Och Teknologier, Tech. Rep., 2010.
[26] N. Kumar, P. N. Belhumeur, A. Biswas, D. W. Jacobs, W. J. Kress,I. Lopez, and J. V. B. Soares, ‘‘Leafsnap: A computer vision system forautomatic plant species identification,’’ in Proc. 12th Eur. Conf. Comput.Vis. (ECCV), Oct. 2012, pp. 502–516. doi: 10.1007/978-3-642-33709-3_36.
[27] A. Krizhevsky, I. Sutskever, and G. E. Hinton, ‘‘ImageNet classificationwith deep convolutional neural networks,’’ in Proc. Adv. Neural Inf. Pro-cess. Syst., 2012, vol. 25, no. 2, pp. 1097–1105. doi: 10.1145/3065386
.[28] B. Liu, X. Yu, A. Yu, P. Zhang, G. Wan, and R. Wang, ‘‘Deepfew-shot learning for hyperspectral image classification,’’ IEEE Trans.Geosci. Remote Sens., vol. 57, no. 4, pp. 2290–2304, Apr. 2019.doi: 10.1109/TGRS.2018.2872830.
[29] S. Zhang, Y.-K. Lei, and Y.-H. Wu, ‘‘Semi-supervised locally discriminantprojection for classification and recognition,’’ Knowl.-Based Syst., vol. 24,no. 3, pp. 341–346, Mar. 2011. doi: 10.1016/j.knosys.2010.11.002.
[30] L. Longlong, J. M. Garibaldi, and H. Dongjian, ‘‘Leaf classification usingmultiple feature analysis based on semi-supervised clustering,’’ J. Intell.Fuzzy Syst., vol. 29, no. 4, pp. 1465–1477, Oct. 2015. doi: 10.3233/IFS-151626.
[31] T. Chen, S. Lu, and J. Fan, ‘‘SS-HCNN: Semi-supervised hierarchicalconvolutional neural network for image classification,’’ IEEE Trans. ImageProcess., vol. 28, no. 5, pp. 2389–2398, May 2019. doi: 10.1109/TIP.2018.2886758.