[论文翻译]V-Net:Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation
论文下载: 地址
V-Net: Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation
V-Net: 用于三维医学图像分割的全卷积神经网络
Abstract. Convolutional Neural Networks (CNNs) have been recently employed to solve problems from both the computer vision and medical image analysis fields. Despite their popularity, most approaches are only able to process 2D images while most medical data used in clinical practice consists of 3D volumes. In this work we propose an approach to 3D image segmentation based on a volumetric, fully convolutional, neural network. Our CNN is trained end-to-end on MRI volumes depicting prostate, and learns to predict segmentation for the whole volume at once. We introduce a novel objective function, that we optimise during training, based on Dice coefficient. In this way we can deal with situations where there is a strong imbalance between the number of foreground and background voxels. To cope with the limited number of annotated volumes available for training, we augment the data applying random non-linear transformations and histogram matching. We show in our experimental evaluation that our approach achieves good performances on challenging test data while requiring only a fraction of the processing time needed by other previous methods.
摘要卷积神经网络(Convolutional Neural Networks, CNNs)最近被应用于解决计算机视觉和医学图像分析领域的问题。尽管它们很受欢迎,但大多数方法只能处理2D图像,而在临床实践中使用的大多数医疗数据是由3D组成的。在这项工作中,我们提出了一种基于三维全卷积神经网络的三维图像分割方法。我们的CNN可以进行端到端的前列腺核磁共振成像的训练,并能够预测整个三维图像的分割。我们提出了一种新的基于Dice系数的目标函数,并在训练过程中对其进行了优化。这样,我们就可以处理前景和背景体素数量严重不平衡的情况。为了处理有限数量的可用于训练的标签图,我们使用随机非线性转换和直方图匹配来增加数据。我们的实验评估表明,我们的方法在具有挑战性的测试数据上取得了良好的性能,而只需要其他以前的方法的一小部分处理时间。
1 Introduction and Related Work
Recent research in computer vision and pattern recognition has highlighted the capabilities of Convolutional Neural Networks (CNNs) to solve challenging tasks such as classification, segmentation and object detection, achieving state-of-theart performances. This success has been attributed to the ability of CNNs to learn a hierarchical representation of raw input data, without relying on handcrafted features. As the inputs are processed through the network layers, the level of abstraction of the resulting features increases. Shallower layers grasp local information while deeper layers use filters whose receptive fields are much broader that therefore capture global information [19].
最近在计算机视觉和模式识别方面的研究突出了卷积神经网络(CNNs)解决分类、分割和对象检测等挑战性任务的能力,实现了最先进的性能。这一成功归功于CNNs在不依赖手工特性的情况下,能够学习原始输入数据的层次表示。当输入通过网络层进行处理时,结果特性的抽象级别就会增加。较浅的层捕获局部信息,而较深的层使用卷积核,其接受域更广,因此捕获全局信息[19]。
Segmentation is a highly relevant task in medical image analysis. Automatic delineation of organs and structures of interest is often necessary to perform tasks such as visual augmentation [10], computer assisted diagnosis [12], interventions [20] and extraction of quantitative indices from images [1]. In particular, since diagnostic and interventional imagery often consists of 3D images, being able to perform volumetric segmentations by taking into account the whole volume content at once, has a particular relevance. In this work, we aim to segment prostate MRI volumes. This is a challenging task due to the wide range of appearance the prostate can assume in different scans due to deformations and variations of the intensity distribution. Moreover, MRI volumes are often affected by artefacts and distortions due to field inhomogeneity. Prostate segmentation is nevertheless an important task having clinical relevance both during diagnosis, where the volume of the prostate needs to be assessed [13], and during treatment planning, where the estimate of the anatomical boundary needs to be accurate [4,20].
分割是医学图像分析中一个高度相关的课题。自动描绘感兴趣的器官和结构通常是必要的执行任务,如视觉增强[10],计算机辅助诊断[12],干预[20]和提取定量指标的图像[1]。特别是,由于诊断和介入图像通常由三维图像组成,因此能够同时考虑整个三维内容进行体积分段具有特殊的相关性。在这项工作中,我们的目标是分割三维前列腺MRI。这是一个具有挑战性的任务,因为前列腺可以在不同的扫描中呈现广泛的外观,由于变形和强度分布的变化。此外,由于场的不均匀性,磁共振成像经常受到人工制品和畸变的影响。然而,前列腺分割是一项重要的临床任务,在诊断期间,前列腺体需要评估[13],在治疗计划期间,解剖边界的估计需要准确[4,20]。
CNNs have been recently used for medical image segmentation. Early approaches obtain anatomy delineation in images or volumes by performing patchwise image classification. Such segmentations are obtained by only considering local context and therefore are prone to failure, especially in challenging modalities such as ultrasound, where a high number of mis-classified voxel are to be expected. Post-processing approaches such as connected components analysis normally yield no improvement and therefore, more recent works, propose to use the network predictions in combination with Markov random fields [6], voting strategies [9] or more traditional approaches such as level-sets [2]. Patch-wise approaches also suffer from efficiency issues. When densely extracted patches are processed in a CNN, a high number of computations is redundant and therefore the total algorithm runtime is high. In this case, more efficient computational schemes can be adopted.
近年来,CNNs被广泛应用于医学图像分割。早期的方法是通过分块图像分类来获得图像或卷体的解剖轮廓。这种分割只考虑局部环境,因此很容易失败,特别是在具有挑战性的模式,如超声,其中大量错误分类的体素是预期的。后处理方法,如连接成分分析,通常不会产生改善,因此,最近的工作,建议使用网络预测与马尔科夫随机域[6],投票策略[9]或更传统的方法,如水平集[2]相结合。补丁式方法也存在效率问题。当在CNN中处理密集提取的patch时,大量的计算是冗余的,因此总的算法运行时是高的。在这种情况下,可以采用更有效的计算方案。
Fully convolutional network trained end-to-end were so far applied only to 2D images both in computer vision [11,8] and microscopy image analysis [14]. These models, which served as an inspiration for our work, employed different network architectures and were trained to predict a segmentation mask, delineating the structures of interest, for the whole image. In [11] a pre-trained VGG network architecture [15] was used in conjunction with its mirrored, de-convolutional, equivalent to segment RGB images by leveraging the descriptive power of the features extracted by the innermost layer. In [8] three fully convolutional deep neural networks, pre-trained on a classification task, were refined to produce segmentations while in [14] a brand new CNN model, especially tailored to tackle biomedical image analysis problems in 2D, was proposed.
到目前为止,无论是在计算机视觉[11,8]还是在显微镜图像分析[14]中,端到端的全卷积网络都只适用于二维图像。这些模型为我们的工作提供了灵感,它们使用了不同的网络架构,并经过训练来预测整个图像的分割掩模,描述感兴趣的结构。在[11]中,一个预先训练的VGG网络架构[15]与它的镜像、去卷积,通过利用最内层提取的特征的描述能力来对RGB图像进行分割。在[8]中,我们对3个完全卷积的深度神经网络进行了分类任务的训练,并对其进行了细化以产生分段,而在[14]中,我们提出了一个全新的CNN模型,特别针对二维生物医学图像分析问题。
In this work we present our approach to medical image segmentation that leverages the power of a fully convolutional neural networks, trained end-to-end, to process MRI volumes. Differently from other recent approaches we refrain from processing the input volumes slice-wise and we propose to use volumetric convolutions instead. We propose a novel objective function based on Dice coefficient maximisation, that we optimise during training. We demonstrate fast and accurate results on prostate MRI test volumes and we provide direct comparison with other methods which were evaluated on the same test data
在这项工作中,我们提出了我们的方法,医学图像分割,利用完全卷积神经网络的力量,训练端到端,以处理三维核磁共振。与最近的其他方法不同的是,我们避免了用切片的方式处理,而是使用三维卷积。我们提出了一个新的目标函数基于dice系数最大化,我们优化训练。我们实验证明了能够在三维前列腺MRI的测试中获得快速和准确的结果,我们提供了直接的比较与其他方法的评估在相同的测试数据
2 Method
Fig. 2. Schematic representation of our network architecture. Our custom implementation of Caffe [5] processes 3D data by performing volumetric convolutions. Best viewed in electronic format.
图2所示。我们的网络结构的示意图。我们的Caffe[5]的自定义实现通过卷积来处理3D数据。最好以电子格式观看。
In Figure 2 we provide a schematic representation of our convolutional neural network. We perform convolutions aiming to both extract features from the data and, at the end of each stage, to reduce its resolution by using appropriate stride. The left part of the network consists of a compression path, while the right part decompresses the signal until its original size is reached. Convolutions are all applied with appropriate padding.
在图2中,我们提供了卷积神经网络的示意图。我们执行卷积的目的是从数据中提取特征,并在每个阶段的末尾使用适当的stride来降低其分辨率。网络的左侧部分由压缩路径组成,而右侧部分对信号进行解压,直到达到其原始大小。卷积都使用适当的填充。
The left side of the network is divided in different stages that operate at different resolutions. Each stage comprises one to three convolutional layers. Similarly to the approach presented in [3], we formulate each stage such that it learns a residual function: the input of each stage is (a) used in the convolutional layers and processed through the non-linearities and (b) added to the output of the last convolutional layer of that stage in order to enable learning a residual function. As confirmed by our empirical observations, this architecture ensures convergence in a fraction of the time required by a similar network that does not learn residual functions.
网络的左侧被划分为不同的阶段,以不同的分辨率运行。每个尺度包括一到三个卷积层。类似于[3]中提出的方法,我们计算每个尺度使其学习剩余功能:输入的每个阶段(a)中使用卷积层和处理非线性和(b)添加到输出的最后卷积层阶段,为了使学习一个剩余函数。正如我们的经验观察所证实的那样,这种架构确保了一小部分时间内收敛,相比于那些不学习剩余函数的类似网络。
The convolutions performed in each stage use volumetric kernels having size 5×5×5 voxels. As the data proceeds through different stages along the compression path, its resolution is reduced. This is performed through convolution with 2 × 2 × 2 voxels wide kernels applied with stride 2 (Figure 3). Since the second operation extracts features by considering only non overlapping 2×2×2 volume patches, the size of the resulting feature maps is halved. This strategy serves a similar purpose as pooling layers that, motivated by [16] and other works discouraging the use of max-pooling operations in CNNs, have been replaced in our approach by convolutional ones. Moreover, since the number of feature channels doubles at each stage of the compression path of the V-Net, and due to the formulation of the model as a residual network, we resort to these convolution operations to double the number of feature maps as we reduce their resolution. PReLu non linearities are applied throughout the network.
每个阶段的卷积使用体积为5×5×5的卷积核。当数据沿着压缩路径经过不同的阶段时,其分辨率会降低。这是通过与使用stride 2的2×2×2的体素宽核进行卷积来实现的(图3)。由于第二次操作只考虑不重叠的2×2×2的体积patch来提取特征,因此得到的特征图的大小减少了一半。这种策略的作用类似于合用层,在[16]和其他工作的激励下,不鼓励在CNNs中使用最大合用操作,而在我们的方法中,这一层已经被卷积层所取代。此外,由于V-Net压缩路径的每一阶段特征通道的数量都增加了一倍,并且由于模型是一个残差网络,我们通过这些卷积操作来减少特征映射的分辨率,从而使特征映射的数量增加一倍。PReLu非线性应用于整个网络。
Replacing pooling operations with convolutional ones results also to networks that, depending on the specific implementation, can have a smaller memory footprint during training, due to the fact that no switches mapping the output of pooling layers back to their inputs are needed for back-propagation, and that can be better understood and analysed [19] by applying only de-convolutions instead of un-pooling operations.
更换池操作与卷积的结果也网络,根据特定的实现,可以有一个更小的内存空间在训练期间,由于没有开关的输出映射池层反向传播,需要回到他们的输入,可以更好地理解和分析[19]通过应用只有de-convolutions代替un-pooling操作。
Downsampling allows us to reduce the size of the signal presented as input and to increase the receptive field of the features being computed in subsequent network layers. Each of the stages of the left part of the network, computes a number of features which is two times higher than the one of the previous layer.
下行采样允许我们减小作为输入的信号的大小,并增加后续网络层中计算的特征的接受域。网络左侧的每个阶段,计算出的特征数是前一层的两倍。
The right portion of the network extracts features and expands the spatial support of the lower resolution feature maps in order to gather and assemble the necessary information to output a two channel volumetric segmentation. The two features maps computed by the very last convolutional layer, having 1×1×1 kernel size and producing outputs of the same size as the input volume, are converted to probabilistic segmentations of the foreground and background regions by applying soft-max voxelwise. After each stage of the right portion of the CNN, a de-convolution operation is employed in order increase the size of the inputs (Figure 3) followed by one to three convolutional layers involving half the number of 5 × 5 × 5 kernels employed in the previous layer. Similar to the left part of the network, also in this case we resort to learn residual functions in the convolutional stages.
网络的右部提取特征并扩展低分辨率地形图的空间支持,以收集和装配必要的信息,输出两个通道的体积分割。最后一个卷积层的核大小为1×1×1,输出大小与输入体积相同,通过应用软max voxelwise将这两个特征映射转化为前景和背景区域的概率分割。在CNN右侧的每一阶段之后,为了增加输入的大小,需要进行去卷积操作(图3),然后是1到3个卷积层,这些卷积层的数量是上一层的5×5×5个内核数量的一半。与网络的左侧部分类似,在这种情况下,我们还需要学习卷积阶段的剩余函数。
Fig. 3. Convolutions with appropriate stride can be used to reduce the size of the data. Conversely, de-convolutions increase the data size by projecting each input voxel to a bigger region through the kernel
图3所示。使用适当的步幅进行卷积可以减少数据的大小。相反,去卷积通过内核将每个输入体素投射到更大的区域,从而增加了数据的大小
Similarly to [14], we forward the features extracted from early stages of the left part of the CNN to the right part. This is schematically represented in Figure 2 by horizontal connections. In this way we gather fine grained detail that would be otherwise lost in the compression path and we improve the quality of the final contour prediction. We also observed that when these connections improve the convergence time of the model.
与[14]类似,我们将从CNN左侧早期提取的特征转发到右侧。这在图2中通过水平连接表示。通过这种方式,我们收集了在压缩路径中可能丢失的细粒度细节,并提高了最终轮廓预测的质量。我们还观察到,当这些连接提高了模型的收敛时间。
We report in Table 1 the receptive fields of each network layer, showing the fact that the innermost portion of our CNN already captures the content of the whole input volume. We believe that this characteristic is important during segmentation of poorly visible anatomy: the features computed in the deepest layer perceive the whole anatomy of interest at once, since they are computed from data having a spatial support much larger than the typical size of the anatomy we seek to delineate, and therefore impose global constraints.
我们在表1中报告了每个网络层的接受域,显示CNN的最内层已经捕获了整个输入卷的内容。我们相信这一特点是重要的在不可见的解剖学细分:最深的特性计算层感知整个解剖学的兴趣,因为它们是计算从数据空间的支持远远大于我们所寻求的解剖学的典型尺寸描述,因此对全球约束。
3 Dice loss layer
The network predictions, which consist of two volumes having the same resolution as the original input data, are processed through a soft-max layer which outputs the probability of each voxel to belong to foreground and to background. In medical volumes such as the ones we are processing in this work, it is not uncommon that the anatomy of interest occupies only a very small region of the scan. This often causes the learning process to get trapped in local minima of the loss function yielding a network whose predictions are strongly biased towards background. As a result the foreground region is often missing or only partially detected. Several previous approaches resorted to loss functions based on sample re-weighting where foreground regions are given more importance than background ones during learning. In this work we propose a novel objective function based on dice coefficient, which is a quantity ranging between 0 and 1 which we aim to maximise. The dice coefficient D between two binary volumes can be written as
网络预测由两卷具有与原始输入数据相同分辨率的数据组成,通过soft-max层处理后,然后输出每个像素属于前景和背景的概率。在医学图像中,如我们正在处理的工作,这是很常见的解剖兴趣只占一个非常小的区域的扫描。这常常导致学习过程陷入局部损失函数的最小值,从而产生一个预测严重偏向于背景的网络。因此,前景区域经常丢失或只被部分检测到。之前的几种方法都采用了基于样本加权的损失函数,在学习过程中,前景区域被赋予了比背景区域更大的重要性。在这项工作中,我们提出了一个新的目标函数基于dice系数,这是一个值在0和1之间,我们的目标是最大化。预测和真实图(像素值为0或1)之间的dice系数D可以表示为
computed with respect to the j-th voxel of the prediction. Using this formulation we do not need to assign weights to samples of different classes to establish the right balance between foreground and background voxels, and we obtain results that we experimentally observed are much better than the ones computed through the same network trained optimising a multinomial logistic loss with sample re-weighting (Fig. 6).
根据预测的第j个体素进行计算。使用这个公式我们不需要分配权重的不同类别样本建立前景和背景像素点之间的平衡,在相同的网络中用dice loss的效果要比多项logistic loss好很多(图6)。
3.1 Training
Our CNN is trained end-to-end on a dataset of prostate scans in MRI. An example of the typical content of such volumes is shown in Figure 1. All the volumes processed by the network have fixed size of 128 × 128 × 64 voxels and a spatial resolution of 1 × 1 × 1:5 millimeters.
我们的CNN是在核磁共振的前列腺扫描数据集上端对端训练的。此类卷的典型内容的示例如图1所示。网络处理的所有体块大小固定为128×128×64体块,空间分辨率为1×1×1:5毫米。
Annotated medical volumes are not easy to obtain due to the fact that one or more experts are required to manually trace a reliable ground truth annotation and that there is a cost associated with their acquisition. In this work we found necessary to augment the original training dataset in order to obtain robustness and increased precision on the test dataset
由于需要一名或多名专家手动跟踪可靠的地面真相注释,并且获取这些注释需要花费一定的成本,因此很难获得带注释的医疗卷。在这项工作中,我们发现有必要扩大原始训练数据集,以获得鲁棒性和增加测试数据集的精度
During every training iteration, we fed as input to the network randomly deformed versions of the training images by using a dense deformation field obtained through a 2 × 2 × 2 grid of control-points and B-spline interpolation. This augmentation has been performed "on-the-fly", prior to each optimisation iteration, in order to alleviate the otherwise excessive storage requirements. Additionally we vary the intensity distribution of the data by adapting, using histogram matching, the intensity distributions of the training volumes used in each iteration, to the ones of other randomly chosen scans belonging to the dataset.
在每次训练迭代过程中,我们利用2×2×2控制点网格和b样条插值得到的密集变形场,将随机变形的训练图像作为输入输入到网络中。为了缓解过多的存储需求,在每次优化迭代之前,都会“动态地”执行这种扩展。此外,我们还通过使用直方图匹配来改变数据的强度分布,即每次迭代中使用的训练卷的强度分布,以适应属于数据集的其他随机选择的扫描的强度分布。
3.2 Testing
A Previously unseen MRI volume can be segmented by processing it in a feedforward manner through the network. The output of the last convolutional layer, after soft-max, consists of a probability map for background and foreground. The voxels having higher probability (> 0:5) to belong to the foreground than to the background are considered part of the anatomy
以前未见过的核磁共振体积可以通过网络前馈的方式进行分割。最后一个卷积层的输出,经过soft-max,由一个背景和前景的概率图组成。体素有更高的可能性(> 0:5)属于前景比背景被认为是解剖学的一部分
4 Results
We trained our method on 50 MRI volumes, and the relative manual ground truth annotation, obtained from the "PROMISE2012" challenge dataset [7]. This dataset contains medical data acquired in different hospitals, using different equipment and different acquisition protocols. The data in this dataset is representative of the clinical variability and challenges encountered in clinical settings. As previously stated we massively augmented this dataset through random transformation performed in each training iteration, for each mini-batch fed to the network. The mini-batches used in our implementation contained two volumes each, mainly due to the high memory requirement of the model during training. We used a momentum of 0:99 and a initial learning rate of 0:0001 which decreases by one order of magnitude every 25K iterations.
我们从“承诺2012”挑战数据集[7]中获得了50个MRI卷和相关的手动地面真相注释,并对我们的方法进行了训练。该数据集包含不同医院使用不同设备和不同采集协议获取的医疗数据。该数据集中的数据代表了在临床环境中遇到的临床变异性和挑战。如前所述,我们通过在每个训练迭代中执行的随机转换,为每个提供给网络的迷你批处理大量扩充了这个数据集。在我们的实现中使用的小批包含两个卷,主要是由于在培训期间模型的高内存需求。我们使用了0:99的动量和0:0001的初始学习率,每25K迭代减少一个数量级。
We tested V-Net on 30 MRI volumes depicting prostate whose ground truth annotation was secret. All the results reported in this section of the paper were obtained directly from the organisers of the challenge after submitting the segmentation obtained through our approach. The test set was representative of the clinical variability encountered in prostate scans in real clinical settings [7].
我们使用V-Net测试了30个MRI卷,这些卷描绘了前列腺的地物真实注释是秘密的。所有报告的结果在这部分的论文是直接从主办单位的挑战后,提交分割通过我们的方法获得。该测试集代表了在真实的临床环境中前列腺扫描所遇到的临床变异性。
We evaluated the approach performance in terms of Dice coefficient, Hausdorff distance of the predicted delineation to the ground truth annotation and in terms of score obtained on the challenge data as computed by the organisers of "PROMISE 2012" [7]. The results are shown in Table 2 and Fig. 5.
我们通过Dice系数、预测轮廓到地面真值注释的Hausdorff距离以及“PROMISE 2012”[7]组织者计算的挑战数据得分来评估该方法的性能。结果如表2和图5所示。
Our implementation5 was realised in python, using a custom version of the Caffe6 [5] framework which was enabled to perform volumetric convolutions via CuDNN v3. All the trainings and experiments were ran on a standard workstation equipped with 64 GB of memory, an Intel(R) Core(TM) i7-5820K CPU working at 3.30GHz, and a NVidia GTX 1080 with 8 GB of video memory. We let our model train for 48 hours, or 30K iterations circa, and we were able to segment a previously unseen volume in circa 1 second. The datasets were first normalised using the N4 bias filed correction function of the ANTs framework [17] and then resampled to a common resolution of 1 × 1 × 1:5 mm. We applied random deformations to the scans used for training by varying the position of the control points with random quantities obtained from gaussian distribution with zero mean and 15 voxels standard deviation. Qualitative results can be seen in Fig. 4.
我们的实现是在python中实现的,使用的是自定义版本的咖啡因6[5]框架,它能够通过CuDNN v3执行卷。所有的训练和实验都是在一个标准的工作站进行的,该工作站配备了64 GB的内存,一个Intel(R) Core(TM) i7-5820K CPU,工作频率为3.30GHz,以及一个NVidia GTX 1080,带有8 GB的视频内存。我们让我们的模型训练48小时,或者大约30K的迭代,并且我们能够在大约1秒的时间内分割出一个以前未见过的卷。首先利用蚂蚁框架[17]的N4偏场校正函数对数据集进行归一化处理,然后将其重采样到1×1×1:5 mm的共同分辨率。我们将随机变形应用到用于训练的扫描中,通过改变控制点的位置来获得随机数量,这些随机数量来自于零均值和15个体素标准偏差的高斯分布。定性结果如图4所示。
5 Conclusion
We presented and approach based on a volumetric convolutional neural network that performs segmentation of MRI prostate volumes in a fast and accurate manner. We introduced a novel objective function that we optimise during training based on the Dice overlap coefficient between the predicted segmentation and the ground truth annotation. Our Dice loss layer does not need sample re-weighting when the amount of background and foreground pixels is strongly unbalanced and is indicated for binary segmentation tasks. Although we inspired our architecture to the one proposed in [14], we divided it into stages that learn residuals and, as empirically observed, improve both results and convergence time. Future works will aim at segmenting volumes containing multiple regions in other modalities such as ultrasound and at higher resolutions by splitting the network over multiple GPUs
我们提出了一种基于体积卷积神经网络的MRI前列腺体积快速准确分割方法。我们提出了一种新的目标函数,在训练过程中根据预测分割和地面真值注释之间的骰子重叠系数进行优化。我们的骰子损失层不需要样本重新加权时,背景和前景像素的数量是强烈不平衡的,并表示为二值分割任务。虽然我们的架构灵感来自于[14]中提出的架构,但是我们将其划分为几个阶段,这些阶段学习残差,并且根据经验观察,提高了结果和收敛时间。未来的工作将致力于将包含多个区域的体块以其他形式(如超声)分割,并通过在多个gpu上分割网络来实现更高的分辨率
6 Acknowledgement
We would like to acknowledge NVidia corporation, that donated a Tesla K40 GPU to our group enabling this research, Dr. Geert Litjens who dedicated some of his time to evaluate our results against the ground truth of the PROMISE 2012 dataset and Ms. Iro Laina for her support to this project
我们愿意承认NVidia公司捐赠的特斯拉K40 GPU来我们组启用这个研究,博士Geert Litjens他专用的一些时间来评估我们的结果对承诺的地面实况2012数据集和女士——Laina支持这个项目
![[论文翻译]V-Net:Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation_第1张图片](http://img.e-com-net.com/image/info8/3aeb5e25e20b46c08bcb9d2e31195ab4.jpg)
![[论文翻译]V-Net:Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation_第2张图片](http://img.e-com-net.com/image/info8/e68e1881aff94cd6a35b3209d7694a8c.jpg)
![[论文翻译]V-Net:Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation_第3张图片](http://img.e-com-net.com/image/info8/3f826dd6a63d413395b7e61b23dd072e.jpg)
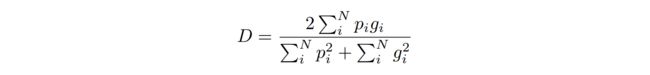
![[论文翻译]V-Net:Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation_第4张图片](http://img.e-com-net.com/image/info8/4f49a6a0588e4808b71223b619b4002d.jpg)
![[论文翻译]V-Net:Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation_第5张图片](http://img.e-com-net.com/image/info8/c865771bde234a2db2acd8252c97b7f2.jpg)
![[论文翻译]V-Net:Fully Convolutional Neural Networks for Volumetric Medical Image Segmentation_第6张图片](http://img.e-com-net.com/image/info8/25e1a55bff99494c884b8388078ff093.jpg)