linux 安装 python3.6 pip3 setuptools并用python解析dcm影像文件
一、安装 python3.6
1.安装依赖环境
yum -y install zlib-devel bzip2-devel openssl-devel ncurses-devel sqlite-devel readline-devel tk-devel gdbm-devel db4-devel libpcap-devel xz-devel
2.下载 Python3
wget https://www.python.org/ftp/python/3.6.1/Python-3.6.1.tgz
3.安装 python3
(1)创建文件夹
mkdir -p /home/mpx/python3
我是解压下载好的 Python-3.x.x.tgz 包(具体包名因你下载的 Python 具体版本不同⽽不同,如:我下载的是Python3.6.1那我这里就是 Python-3.6.1.tgz)

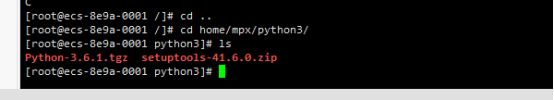
(2)解压
tar -zxvf Python-3.6.1.tgz
4.进入解压后的目录,编译安装
cd Python-3.6.1
./configure --prefix=/home/mpx/python3
如果遇上如下报错

由于没有gcc导致的,安装gcc即可解决:
yum install gcc
没有报错或者安装完gcc 向下继续
make
make install
或者
make && make install
5.建立 python3 的软链
ln -s /home/mpx/python3/bin/python3 /usr/bin/python3
6.将/usr/local/python3/bin加入 PATH
vim ~/.bash_profile
# .bash_profile
# Get the aliases and functions
if [ -f ~/.bashrc ]; then
. ~/.bashrc
fi
# User specific environment and startup programs
PATH=$PATH:$HOME/bin:/home/mpx/python3/bin
export PATH
使配置文件生效
source ~/.bash_profile
校验是否安装成功 分别执行一下
python3 -V
pip3 -V
7.不行的话在创建一下 pip3 的软链接(我也不清楚这一步有什么用)
ln -s /usr/local/python3/bin/pip3 /usr/bin/pip3
二、安装 pip 前需要前置安装setuptools 到官网下载
1.进行解压
unzip setuptools-41.6.0.zip
2.进入目录
cd setuptools-41.6.0/
编译
python3 setup.py build
安装
python3 setup.py install
安装pip
easy_install pip
pip3 install SimpleITK -i https://pypi.tuna.tsinghua.edu.cn/simple
pip3 install numpy -i https://pypi.tuna.tsinghua.edu.cn/simple
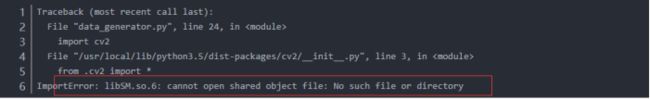
pip3 install opencv-python -i https://pypi.tuna.tsinghua.edu.cn/simple
pip3 install pydicom -i https://pypi.tuna.tsinghua.edu.cn/simple
sudo yum install -y python-qt4
usr下创建python文件夹并上传一个dcm文件
mkdir /usr/python
python将dcm源文件 转图片代码
import SimpleITK as sitk
import numpy as np
import cv2
import sys
def coverJPG(inputfile, outputfile):
ds_array = sitk.ReadImage(inputfile) # 读取dicom文件的相关信息
img_array = sitk.GetArrayFromImage(ds_array) # 获取array
# SimpleITK读取的图像数据的坐标顺序为zyx,即从多少张切片到单张切片的宽和高,此处我们读取单张,因此img_array的shape
# 类似于 (1,height,width)的形式
shape = img_array.shape
img_array = np.reshape(img_array, (shape[1], shape[2])) # 获取array中的height和width
low = np.min(img_array)
high = np.max(img_array)
lungwin = np.array([low * 1., high * 1.])
newimg = (img_array - lungwin[0]) / (lungwin[1] - lungwin[0]) # 归一化
newimg = (newimg * 255).astype('uint8') # 将像素值扩展到[0,255]
cv2.imwrite(outputfile, newimg, [int(cv2.IMWRITE_JPEG_QUALITY), 100])
print('FINISHED')
if __name__ == "__main__":
coverJPG(sys.argv[1], sys.argv[2])
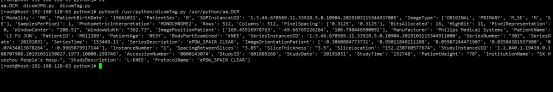
测试转换图片是否成功
python3 /usr/python/dicomJPG.py /usr/python/aa.DCM /usr/python/bb.jpg
import pydicom
import sys
def loadFileInformation(filename):
information = {}
ds = pydicom.read_file(filename)
if(hasattr(ds, 'Modality')):
information['Modality'] = ds.Modality
if(hasattr(ds, 'PatientBirthDate')):
information['PatientBirthDate'] = ds.PatientBirthDate
if(hasattr(ds, 'PatientSex')):
information['PatientSex'] = ds.PatientSex
if(hasattr(ds, 'SOPInstanceUID')):
information['SOPInstanceUID'] = ds.SOPInstanceUID
if(hasattr(ds, 'ImageType')):
information['ImageType'] = ds.ImageType
if(hasattr(ds, 'SamplesPerPixel')):
information['SamplesPerPixel'] = ds.SamplesPerPixel
if(hasattr(ds, 'PhotometricInterpretation')):
information['PhotometricInterpretation'] = ds.PhotometricInterpretation
if(hasattr(ds, 'Rows')):
information['Rows'] = ds.Rows
if(hasattr(ds, 'Columns')):
information['Columns'] = ds.Columns
if(hasattr(ds, 'PixelSpacing')):
information['PixelSpacing'] = ds.PixelSpacing
if(hasattr(ds, 'BitsAllocated')):
information['BitsAllocated'] = ds.BitsAllocated
if(hasattr(ds, 'HighBit')):
information['HighBit'] = ds.HighBit
if(hasattr(ds, 'PixelRepresentation')):
information['PixelRepresentation'] = ds.PixelRepresentation
if(hasattr(ds, 'WindowCenter')):
information['WindowCenter'] = ds.WindowCenter
if(hasattr(ds, 'WindowWidth')):
information['WindowWidth'] = ds.WindowWidth
if(hasattr(ds, 'RescaleIntercept')):
information['RescaleIntercept'] = ds.RescaleIntercept
if(hasattr(ds, 'RescaleSlope')):
information['RescaleSlope'] = ds.RescaleSlope
if(hasattr(ds, 'RescaleType')):
information['RescaleType'] = ds.RescaleType
if(hasattr(ds, 'ImagePositionPatient')):
information['ImagePositionPatient'] = ds.ImagePositionPatient
if(hasattr(ds, 'Manufacturer')):
information['Manufacturer'] = ds.Manufacturer
if(hasattr(ds, 'PatientName')):
information["PatientName"] = ds.PatientName
if(hasattr(ds, 'PatientID')):
information["PatientID"] = ds.PatientID
if(hasattr(ds, 'PatientAge')):
information["PatientAge"] = ds.PatientAge
if(hasattr(ds, 'BodyPartExamined')):
information["BodyPartExamined"] = ds.BodyPartExamined
if(hasattr(ds, 'SeriesInstanceUID')):
information["SeriesInstanceUID"] = ds.SeriesInstanceUID
if(hasattr(ds, 'SeriesNumber')):
information["SeriesNumber"] = ds.SeriesNumber
if(hasattr(ds, 'SeriesDate')):
information["SeriesDate"] = ds.SeriesDate
if(hasattr(ds, 'SeriesTime')):
information["SeriesTime"] = ds.SeriesTime
if(hasattr(ds, 'SeriesDescription')):
information["SeriesDescription"] = ds.SeriesDescription
if(hasattr(ds, 'ImageOrientationPatient')):
information["ImageOrientationPatient"] = ds.ImageOrientationPatient
if(hasattr(ds, 'InstanceNumber')):
information["InstanceNumber"] = ds.InstanceNumber
if(hasattr(ds, 'SpacingBetweenSlices')):
information["SpacingBetweenSlices"] = ds.SpacingBetweenSlices
if(hasattr(ds, 'SliceThickness')):
information["SliceThickness"] = ds.SliceThickness
if(hasattr(ds, 'SliceLocation')):
information["SliceLocation"] = ds.SliceLocation
if(hasattr(ds, 'StudyInstanceUID')):
information["StudyInstanceUID"] = ds.StudyInstanceUID
if(hasattr(ds, 'AccessionNumber')):
information["AccessionNumber"] = ds.AccessionNumber
if(hasattr(ds, 'StudyID')):
information["StudyID"] = ds.StudyID
if(hasattr(ds, 'StudyDate')):
information["StudyDate"] = ds.StudyDate
if(hasattr(ds, 'StudyTime')):
information["StudyTime"] = ds.StudyTime
if(hasattr(ds, 'PatientWeight')):
information["PatientWeight"] = ds.PatientWeight
if(hasattr(ds, 'InstitutionName')):
information["InstitutionName"] = ds.InstitutionName
if(hasattr(ds, 'ModalitiesInStudy')):
information["ModalitiesInStudy"] = ds.ModalitiesInStudy
if(hasattr(ds, 'StudyDescription')):
information["StudyDescription"] = ds.StudyDescription
if(hasattr(ds, 'ProtocolName')):
information["ProtocolName"] = ds.ProtocolName
# print (ds.dir())
#
# print (ds)
print (information)
return information
# a=loadFileInformation(path)
# #
# # print (a)
if __name__ == "__main__":
loadFileInformation(sys.argv[1]);