数据导入
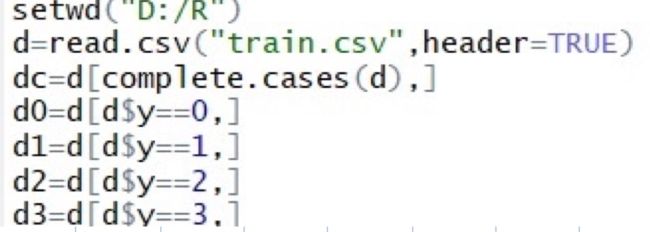
setwd("D:/R")
或者直接getwd()放入学校路径
A=read.csv('',header=TRUE) 有表头true 无 false
数据清理
#1.空值
data=data[complete.cases(data),]#去空值
data=data[!complete.cases(wine),]#显示空值
#2.去重复值
data=unique(data)
#3.查看缺失值
c=is.na(data)
#4.标记缺失值
data() 列出已载入的包中的所有数据集。
data(package = .packages(all.available = TRUE)) 列出已安装的包中的所有数据集。
y = rep(c(1, 2, 3), c(20, 20, 20))
生成20个1 20个2 20个3
y=c(rep(-1,10),rep(1,10))
rep 重复函数 -1 重复出现十次
rnorm()函数产生一系列的随机数,随机数个数,均值和标准差都可以设定
sample 有无放回生成随机数 https://blog.csdn.net/Heidlyn/article/details/56013509
cor() 函数计算两两变量之间的相关系数的矩阵
数据中心化: scale(data,center=T,scale=F)
数据标准化: scale(data,center=T,scale=T) 或默认参数scale(data)
进行pca之前一般先变量标准化
决策树 分类树 剪枝条
决策树(https://blog.csdn.net/u010089444/article/details/53241218)
ID3算法 选择信息增益最大的方向进行分支标准
https://blog.csdn.net/xiaohukun/article/details/78055132
信息增益: 信息熵-条件熵
在决策树算法的学习过程中,信息增益是特征选择的一个重要指标,它定义为一个特征能够为分类系统带来多少信息,带来的信息越多,说明该特征越重要,相应的信息增益也就越大。
https://www.zhihu.com/question/22104055
信息熵越大说明事件的无序程度越高
信息熵越小说明事件的有序程度越高
https://blog.csdn.net/wxn704414736/article/details/80512705
CART
gini越小 越纯
最小的切分点最为最优切分点 使用该切分点将数据切分为两个子集https://blog.csdn.net/wsp_1138886114/article/details/80955528
生成树枝+剪枝
https://www.cnblogs.com/karlpearson/p/6224148.html
监督学习 【分类 回归 支持向量机】
非监督学习【聚类 主成分】
https://blog.csdn.net/chenKFKevin/article/details/70547549
无监督学习:仅有x值 来
两种主要类型无监督学习:聚类分析,主成分分析
定性的响应变量,定性变量也称为分类变量。
线性回归的因变量(Y)是连续变量,自变量(X)可以是连续变量,也可以是分类变量
logistic 回归与线性回归恰好相反,因变量一定要是分类变量,不可能是连续变量。分类变量既可以是二分类,也可以是多分类,多分类中既可以是有序,也可以是无序。
最小二乘法(https://www.zhihu.com/question/37031188)
竖直投影下来 计算(y-ybar)^2最小
pca 降维工具
协方差矩阵——PCA实现的关键
cov() 计算协方差in R
https://www.zhihu.com/question/41120789
pinkyjie.com/2011/02/24/covariance-pca/
prcomp(data,scale=TRUE) scale对数据进行标准化处理
prcomp pca主成分分析函数
混淆矩阵
https://www.zhihu.com/question/36883196
马氏距离。ROC曲线
蒙特卡洛仿真
支持向量机 (文本分类问题)
https://www.zhihu.com/question/21094489
knn
kmeans
https://zhuanlan.zhihu.com/p/31580379
Version:1.0StartHTML:0000000107EndHTML:0000000974StartFragment:0000000127EndFragment:0000000956
X=rbind(matrix(rnorm(20*50,mean = 0),nrow = 20),matrix(rnorm(20*50,mean = 0.7),nrow = 20),matrix(rnorm(20*50,mean = 1.4),nrow = 20))
X.pca=prcomp(X)
plot(X.pca[,1:2],col=c(rep(1,20),rep(2,20),rep(3,20)))
res=kmeans(X,centers=3)
true_class=c(rep(1,20),rep(2,20),rep(3,30))
table(res$cluster,true_class)
wine.case
https://www.kaggle.com/xvivancos/tutorial-clustering-wines-with-k-means
https://www.kaggle.com/maitree/wine-quality-selection
cov_sdc=cov(wine)
eigen(cov_sdc)
res.pca <- PCA(wine[,-12], graph = TRUE)
eig.val <- get_eigenvalue(res.pca)
eig.val
#数据导入
wine=read.csv()
wine= read.csv('winequality-white.csv',header=TRUE)
wine=winequality_white
#data cleaning
wine = wine[complete.cases(wine),]
#PCA
library(stringr)
library(FactoMineR)
#绘图
res.pca <- PCA(wine[,-12], graph = TRUE)#delete Y=quality, plot the PCA graph
sdc=scale(wine)
pca.d=prcomp(sdc)
summary(pca.d)
#PCA降维
wine=wine[,-9:-11]
#查看定性变量分布,确定定性变量
hist(wine$quality)
#分类
wine0 = wine[wine$quality==3,]
wine1 = wine[wine$quality==4,]
wine2 = wine[wine$quality==5,]
wine3 = wine[wine$quality==6,]
wine4 = wine[wine$quality==7,]
wine5 = wine[wine$quality==8,]
#抽样
label0= sample(c(1:10),dim(wine0[1]),replace= TRUE)
label1= sample(c(1:10),dim(wine1[1]),replace= TRUE)
label2= sample(c(1:10),dim(wine2[1]),replace= TRUE)
label3= sample(c(1:10),dim(wine3[1]),replace= TRUE)
label4= sample(c(1:10),dim(wine4[1]),replace= TRUE)
label5= sample(c(1:10),dim(wine5[1]),replace= TRUE)
wine0_train = wine0[label0<=5,]
wine0_test = wine0[label0>5,]
wine1_train = wine1[label1<=5,]
wine1_test = wine1[label1>5,]
wine2_train = wine2[label2<=5,]
wine2_test = wine2[label2>5,]
wine3_train = wine3[label3<=5,]
wine3_test = wine3[label3>5,]
wine4_train = wine4[label4<=5,]
wine4_test = wine4[label4>5,]
wine5_train = wine5[label5<=5,]
wine5_test = wine4[label5>5,]
wine_train = rbind(wine0_train,wine1_train,wine2_train,wine3_train,wine4_train,wine5_train)
wine_test = rbind(wine0_test,wine1_test,wine2_test,wine3_test,wine4_test,wine5_test)
跑
library(nnet)
re_log = multinomial(quality~.,data= wine_train)
将数据变为定性变量
wine_train$quality = as.factor(wine_train$quality)
######################################
library(rpart)
library(rattle)
library(rpart.plot)
#########################################
ID3 方法生成树枝(信息增益)
re_id3 <-rpart(quality~.,data=wine_train,method="class", parms=list(split="information"))
plot(re_id3)
########################################
CART 方法生成树枝(基尼系数)
re_CART = rpart(quality~.,data= wine_train,method = "class",parms = list(split="gini"),control=rpart.control(cp=0.000001))
plot(re_CART,main = "CART")
找到复杂度最小的值
min = which.min(re_CART$cptable[,4])
剪枝
re_CART_f = prune(re_CART,cp=re_CART$cptable[min,1])
pred_id3 = predict(re_id3,newdata = wine_test)
pred_CART = predict(re_CART,newdata = wine_test,type="class")
table(wine_test$quality,pred_CART)
wine_train$quality= as.factor(wine_train$quality)
随机森林
•library("randomForest")
•data.index = sample(c(1,2), nrow(heart), replace = T, prob = c(0.7, 0.3))
•train_data =heart[which(data.index == 1),]
•test_data =heart[which(data.index == 2),]
•n<-length(names(train_data))
•rate=c()
网格法
for (i in 1:(n-1))
{
mtry=i
for(j in (1:100))
{
set.seed(1234)
rf_train=randomForest(as.factor(train_data$target)~.,data=train_data,mtry=i,ntree=j)
rate[(i-1)*100+j]=mean(rf_train$err.rate)
}
}
z=which.min(rate)
print(z)
展示重要性
importance<-importance(heart_rf)
barplot(heart_rf$importance[,1],main="Input variable importance measure indicator bar chart")
box()
importance(heart_rf,type=2)
varImpPlot(x=heart_rf,sort=TRUE,n.var=nrow(heart_rf$importance),main="scatterplot") #可视化
hist(treesize(heart_rf))
check model
pred<-predict(heart_rf,newdata=data.test)
pred_out_1<-predict(object=heart_rf,newdata=data.test,type="prob")
table<-table(pred,data.test$target)
sum(diag(table))/sum(table)
plot(margin(iris_rf,data.test$target))
----------------------------------------------------------------别管
wine$quality
linear regression
library(ggplot2) # Data visualization
library(readr) # CSV file I/O, e.g. the read_csv function
library(corrgram)
library(lattice) #required for nearest neighbors
library(FNN) # nearest neighbors techniques
library(pROC) # to make ROC curve
install.packages('corrgram')
library(corrgram)
---------------------------------------------------------------------------------
linear_quality = lm(quality ~ fixed acidity+volatile acidity+citric acid+residual sugar+chlorides+free sulfur dioxide+total sulfur dioxide+density, data=wine)
corrgram(wine, lower.panel=panel.shade, upper.panel=panel.ellipse)
wine$poor <- wine$quality <= 4
wine$okay <- wine$quality == 5 | wine$quality == 6
wine$good <- wine$quality >= 7
head(wine)
summary(wine)
KNN
class_knn10 = knn(train=wine[,1:8], test=wine[,1:8], cl=wine$good, k =10)
class_knn20 = knn(train=wine[,1:8],test=wine[,1:8], cl = wine$good, k=20)
table(wine$good,class_knn10)
table(wine$good,class_knn20)
wine123=winequality_white
wine123$poor <- wine$quality <= 4
wine123$okay <- wine$quality == 5 | wine$quality == 6
wine123$good <- wine$quality >= 7
library(rpart) #for trees
tree1 = rpart(good~ alcohol + sulphates+ pH , data = wine123, method="class")
rpart.plot(tree1)
summary(tree1)
pred1 = predict(tree1,newdata=wine123,type="class")
summary(pred1)
summary(wine123$good)
比较模型的准确度
tree2 = rpart(good~ alcohol + volatile acidity +citric acid+ pH , data = wine123, method="class")
tree2 = rpart(good ~ alcohol + volatile acidity + citric acid + sulphates, data = wine123, method="class")
rpart.plot(tree2)
tree2= rpart(good ~ alcohol + volatile acidity + citric acid + sulphates, data = wine123 ,method='class')
pred2 = predict(tree2,newdata=wine123,type="class")
summary(pred2)
summary(wine123$good)
信息熵计算
LDA
决策树
p187 chp4 power function
p212 chp5 boostrap
p215 chp5 loocv
p431 chp10 kmeans
-------------------------------------------------------------------
一、变量的基本定义和基础操作
1. 数值型变量的赋值
a = 5
2. 向量赋值
x = c(1:6) , c()为生成向量对应的函数
3. 向量中元素的访问
x = c(1:6)
x[3] ,中括号中的数字代表所访问的数值在向量x中的位置。
x[-3],负数的标度表示取补集,即返回向量x中除第3位以外的其他元素。
4. 矩阵的定义
B =matrix(c(1:10),nrow=2,ncol=5,byrow=TRUE)
matrix()未定义矩阵的函数,括号中第一个位置为写入矩阵中的元素,nrow参数位行数,ncol参数位列数,byrow=TRUE,表示数据按行的顺序书写。byrow=FALSE 按照列的顺序书写
不打byrow 按照列来输入
5. 矩阵元素的访问
B[1,] 访问矩阵中的第一行
B[,2] 访问矩阵中的第二列
B[2,1]访问矩阵第二行第一列的元素
B[,2:5]访问矩阵2到5列的元素
B[,-4] 访问矩阵中除第4列的元素
6. 常用统计函数
sum()求括号中对象的各个元素和
mean()求括号中对象元素的均值
max() 求括号中对象元素中的最大值
min() 求括号中对象元素中的最小值
7. 其他矩阵信息的提取
dim(B) 返回矩阵的维度,第一个值为行数,第二个值为列数
dim(B)[1]可访问矩阵的行
dim(B)[2] 可访问矩阵的列数 1 代表行 2代表列
length(B)返回对象的长度,(请自行测试返回值是行还是列)
----------------------------------------------------------------------------------------------
1.
a=5
a
2.向量赋值
x=c(1:6)
x
3.向量访问
x[3]
x[-3]
4.矩阵的定义
B=matrix(c(1:10),nrow=2, ncol=5, byrow=TRUE)
5.矩阵访问
B[1,]
B[,1]
B[1:2,3]
B[-1,]
6.返回矩阵维度,告诉你几行几列
dim(B)
dim(B)[2]
length(B)
7.
setwd("D:/R")
d=read.csv("PRSA_data_2010.1.1-2014.12.31.csv",header=TRUE)
d1=d[!is.na(d$pm2.5),]
d1
8.数据源表头名称的访问
names(d1)
names(d1)[3]
9.数据源的基本信息提取
summary(d1),其中3rd Qu.和1st Qu
是把样本排序。第25%大的和第75%的值
10.线性回归的基本形式
lr = glm(pm2.5~., data=d1)
lr
lr2 =glm(pm2.5~No,data=d1)
lr2 =glm(pm2.5~No+TEMP,data=d1)
lr2
lr2 =glm(pm2.5~No+I(No^3) ,data=d1)
lr2
lr2 =glm(pm2.5~poly(No,5) ,data=d1)
11.训练集与测试集的划分
选择的数据为2014,仅此一年的:train = d1[d.1$year==2014,]
test = d1[d1$year<=2013,]
lr3 =glm(pm2.5~TEMP,data=train)
pre = predict(lr3,newdata=test)
12. 训练集数据的交叉验证
label= sample(c(1:10),dim(train)[1],replace=TRUE)
d3 = cbind(train,label)
lr5 =glm(pm2.5~TEMP,data=d3[d3$label!=1,])
pre2 =predict(lr5,newdata= d3[d3$label==1,])
error=pre2-d3[d3$label==1,]$pm2.5
mse =sum(error^2)/length(pre2)
mse
13.线性回归模型计算的顺序
13.1建立模型(选取数据集,输入x与y值
13.2选取training data 与 testing data
e.g.
train = d1[d1$year==2014,]
test = d1[d1$year<=2013,]
13.3用glm函数建模,建模时所用的数据为训练集中较多的数据
e.glr5 =glm(pm2.5~.,data=d3[d3$label!=1,]) 使用d3中label不为1的数据组为数据源进行回归。
13.4用predict函数进行预测,使用测试集数据。以此得到一个由运行模型所得的y值结果。
e.gpre2 =predict(lr5,newdata= d3[d3$label==1,]) 使用d3中label为1的数据组来产生预测值
13.5用将真实值与模型所得值进行做差,得error。计算mse,算出模型准确度。
14.
plot(mse)
plot(mmse)
15.
mse2 = matrix(rep(0,150),nrow=10)
for (j in 1:15)
{
for(i in 1:10)
{
lr7 = glm(pm2.5~poly(No,j),data=d3[d3$label!=i,])
pre4 = predict(lr7,newdata= d3[d3$label==i,])
error=pre4-d3[d3$label==i,]$pm2.5
mse2[i,j]= sum(error^2)/length(pre4)
}
}
16.红包2
x=matrix(rep(0,pack*number),nrow=pack)
number=10000
bonus=20
pack=40
y=matrix(rep(0,pack*number),nrow=pack)
for(j in 1:number)
{
x[1,j]=max(0.01,round(runif(1,min=0.01,max=bonus/pack*2),2)-0.01)
for(i in 1:38)
{
ulimit=(bonus-sum(x[,j]))/(pack-i)*2
x[i+1,j]=max(0.01,round(runif(1,min=0.01,max=ulimit),2))-0.01
}
x[pack,j]=bonus-sum(x[,j])
}
for(k in 1:number)
{
y