统计学习方法(三)1-least_sqaure_method
目录
1、最小二乘法拟合曲线
2、正则化
3、简单交叉验证
4、样条插值
1、最小二乘法拟合曲线
[第一章]
最小二乘法
根据高斯-马尔科夫定理,在线性回归模型( y = β x + ε y = \beta x+\varepsilon y=βx+ε )中,如果误差 ε \varepsilon ε 满足零均值、等方差、互不相关,则最小二乘法估计的回归系数 β ^ \hat \beta β^ 满足最佳线性无偏估计,即方差最小。
使用最小二乘法拟和曲线
对于数据 ( x i , y i ) ( i = 1 , 2 , 3... , m ) (x_i, y_i)(i=1, 2, 3...,m) (xi,yi)(i=1,2,3...,m)
拟合出函数 y ^ = w ^ x \hat y = \hat w x y^=w^x
有误差,即残差: e i = y i ^ − y i e_i=\hat {y_i}-y_i ei=yi^−yi
残差平方和( ∑ i = 1 m ( y i ^ − y i ) 2 \sum_{i=1}^m (\hat {y_i} - y_i)^2 ∑i=1m(yi^−yi)2 )最小时 y ^ \hat y y^ 和 y 相似度最高,更拟合
存在H(x)为n次的多项式, H ( x ) = w 0 + w 1 x + w 2 x 2 + . . . w n x n = ∑ j = 1 n w j x j H(x)=w_0+w_1x+w_2x^2+...w_nx^n = \sum_{j=1}^n w_jx^j H(x)=w0+w1x+w2x2+...wnxn=∑j=1nwjxj
w ( w 0 , w 1 , w 2 , . . . , w n ) w(w_0,w_1,w_2,...,w_n) w(w0,w1,w2,...,wn)为参数
最小二乘法就是要找到一组 w ( w 0 , w 1 , w 2 , . . . , w n ) w(w_0,w_1,w_2,...,w_n) w(w0,w1,w2,...,wn) 使得 ∑ i = 1 n ( h ( x i ) − y i ) 2 \sum_{i=1}^n(h(x_i)-y_i)^2 ∑i=1n(h(xi)−yi)2 (残差平方和) 最小
即,求 m i n ∑ i = 1 m ( h ( x i ) − y i ) 2 min\sum_{i=1}^m(h(x_i)-y_i)^2 min∑i=1m(h(xi)−yi)2
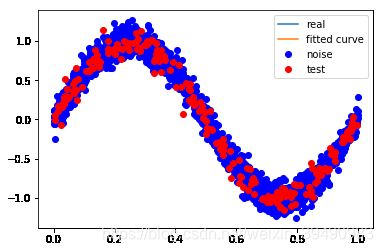
【例1.1 11页】用目标函数 y = s i n 2 π x y=sin2{\pi}x y=sin2πx, 加上一个正太分布的噪音干扰,用多项式去拟合
import numpy as np #数值计算库
import scipy as sp #Scipy是一个高级的科学计算库,它和Numpy联系很密切
from scipy.optimize import leastsq #leastsq()表示最小二乘拟合计算
import matplotlib.pyplot as plt #画图
%matplotlib inline
#%matplotlib inline 可以在Ipython编译器里直接使用,功能是可以内嵌绘图,并且可以省略掉plt.show()这一步。
ps: numpy.poly1d([1,2,3]) 生成 1 x 2 + 2 x 1 + 3 x 0 1x^2+2x^1+3x^0 1x2+2x1+3x0
# 目标函数
def real_func(x):
return np.sin(2*np.pi*x) #定义目标函数real_func
# 多项式
def fit_func(p, x):
f = np.poly1d(p) #定义多项式,p为参数向量,x为随机变量
return f(x)
# 残差
def residuals_func(p, x, y):
ret = fit_func(p, x) - y #定义残差,即多项式拟合值与实际值的差值,y表示输入变量x对应的实际输出变量
return ret
# 十个点
x = np.linspace(0, 1, 10)
x_points = np.linspace(0, 1, 1000)
# 加上正态分布噪音的目标函数的值
y_ = real_func(x)
y = [np.random.normal(0, 0.1)+y1 for y1 in y_]
def fitting1(M=0):
"""
M 为 多项式的最高次数
"""
# 随机初始化多项式参数
p_init = np.random.rand(M+1)
# 最小二乘法
p_lsq = leastsq(residuals_func, p_init, args=(x, y))
# 可视化
plt.plot(x_points, real_func(x_points), label='real')
plt.plot(x_points, fit_func(p_lsq[0], x_points), label='fitted curve')
plt.plot(x, y, 'bo', label='noise')
plt.legend()
return plt.show()
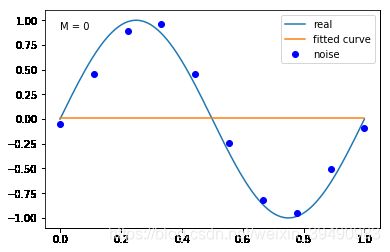
#M=0
plt.text(0,0.9,"M = 0") #加入文本注释
fitting1(M=0)
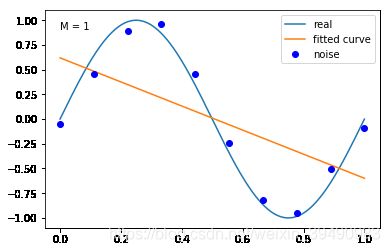
#M=1
plt.text(0,0.9,"M = 1")
fitting1(M=1)
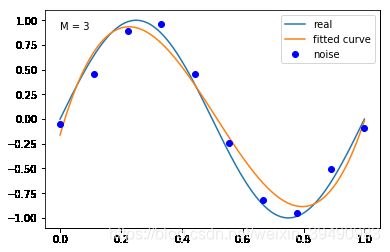
#M=3
plt.text(0,0.9,"M = 3")
fitting1(M=3)
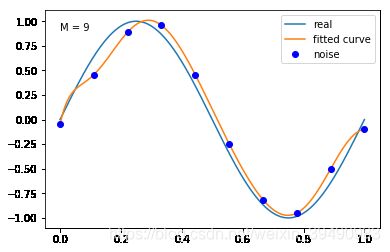
#M=9
plt.text(0,0.9,"M = 9")
fitting1(M=9)
M=3
M=9
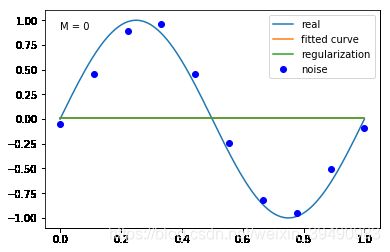
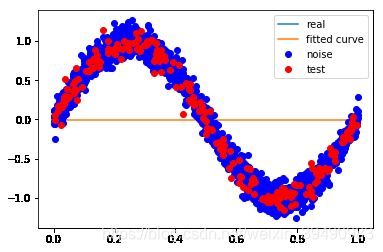
当M=0时,多项式是一条直线 ,数据的拟合效果很差,认为“欠拟合”,欠拟合通常是因为学习能力低下导致的;
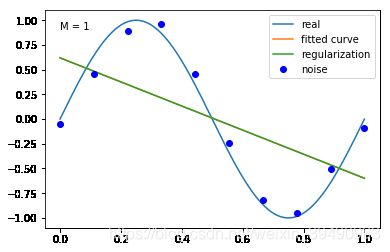
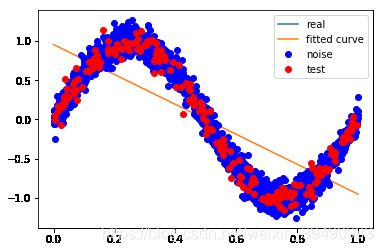
当M=1时,多项式是一条直线 ,数据的拟合效果也很差,认为“欠拟合”;
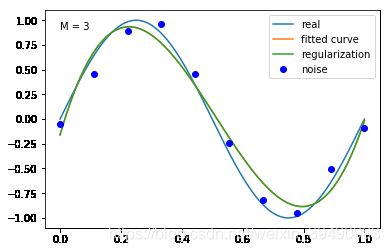
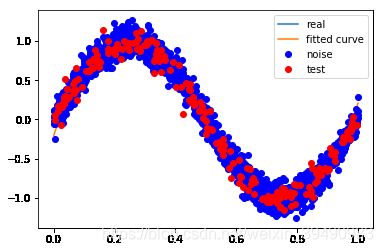
当M=3时,多项式是一条曲线 ,数据的拟合效果比较好,模型也比较简单;
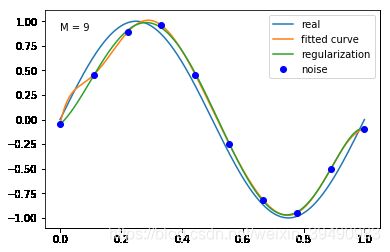
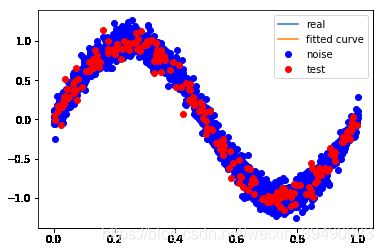
当M=9时,多项式曲线通过了每个数据点,但模型比较复杂,模型对数据预测过好,对未知数据预测很差,认为“过拟合”,过拟合通常是因为所选择模型的复杂度比真模型更高。
对于欠拟合,只需增加模型复杂度即可;
对于过拟合,常用的方法有:正则化与交叉验证。
2、正则化
结果显示过拟合, 引入正则化项(regularizer),降低过拟合
Q ( x ) = ∑ i = 1 n ( h ( x i ) − y i ) 2 + λ ∣ ∣ w ∣ ∣ 2 Q(x)=\sum_{i=1}^n(h(x_i)-y_i)^2+\lambda||w||^2 Q(x)=∑i=1n(h(xi)−yi)2+λ∣∣w∣∣2。
回归问题中,损失函数是平方损失,正则化可以是参数向量的L2范数,也可以是L1范数。
-
L1: regularization*abs( p p p)
-
L2: 0.5 * regularization * np.square( p p p)
正则化的作用是选择经验风险与模型复杂度都较小的模型
正则化符合奥卡姆剃刀原理,在所有可能选择的模型中,能够很好的解释已知数据并且十分简单才是最好的模型。
regularization = 0.0001 #作为调整经验风险和正则化项关系的系数
#加入正则化项的结构风险最小化
def residuals_func_regularization(p, x, y):
ret = fit_func(p, x) - y
ret = np.append(ret, 0.5*regularization*np.sqrt(np.sum(np.square(p)))) # L2范数作为正则化项
return ret
# 最小二乘法,加正则化项
def fitting2(M=0):
"""
M 为 多项式的最高次数
"""
# 随机初始化多项式参数
p_init = np.random.rand(M+1)
# 最小二乘法
p_lsq = leastsq(residuals_func, p_init, args=(x, y))
# 最小二乘法,加入正则化项
p_lsq_regularization = leastsq(residuals_func_regularization, p_init, args=(x, y))
# 可视化
plt.plot(x_points, real_func(x_points), label='real')
plt.plot(x_points, fit_func(p_lsq[0], x_points), label='fitted curve')
plt.plot(x_points, fit_func(p_lsq_regularization[0], x_points), label='regularization')
plt.plot(x, y, 'bo', label='noise')
plt.legend()
return plt.show()
#M=0
plt.text(0,0.9,"M = 0") #加入文本注释
fitting2(M=0)
#M=1
plt.text(0,0.9,"M = 1")
fitting2(M=1)
#M=3
plt.text(0,0.9,"M = 3")
fitting2(M=3)
#M=9
plt.text(0,0.9,"M = 9")
fitting2(M=9)
M = 3
M = 9
加入正则化项后,M = 9多项式的拟合效果更好,过拟合降低
3、简单交叉验证
#简单交叉验证,选择有最小预测误差的模型
from sklearn.model_selection import train_test_split
x = np.linspace(0,1,2000) #创建从0到1,20个等差间隔数列
x_points = np.linspace(0, 1, 1000) #创建从0到1,个数为1000的等差间隔数列
# 加上正态分布随机噪音的目标函数的值
y_ = real_func(x) #将创建的等差数列x作为输入,得到目标函数对应的输出值
y = [np.random.normal(0, 0.1)+y1 for y1 in y_] #解析式,将创建的目标函数的输出值上,随机加入正态分布噪音(其中正态分布的均值为0,标准差为0.1)
test_size = 0.10
seed = 4
x_train,x_test,y_train,y_test = train_test_split(x, y, test_size = test_size, random_state = seed) #random_stste表示随机种子,为了进行可重复的训练,需要固定random_stste
def fitting3(M=0):
"""
M 为 多项式的最高次数
"""
# 随机初始化多项式参数
p_init = np.random.rand(M+1) #来自均匀分布的长度为M+1的随机数,参数表示指定的形状,均匀分布的属于[0, 1)
# 定义最小二乘法,没有加正则化
p_lsq = leastsq(residuals_func, p_init, args=(x_train, y_train)) #来自scipy库的leastsq(),表示最小二乘函数,其中residuals_func为指定的误差函数,p_init为参数初始值,args=(x, y)为误差函数中调用的参数
#leastsq()返回拟合的参数值
print('Fitting Parameters:', p_lsq[0]) #返回拟合的回归系数
# 可视化
plt.plot(x_points, real_func(x_points), label='real')
plt.plot(x_points, fit_func(p_lsq[0], x_points), label='fitted curve') #拟合的M次多项式
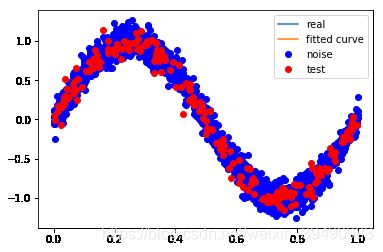
plt.plot(x_train, y_train, 'bo', label='noise')
plt.plot(x_test, y_test, 'ro', label='test')
plt.legend() #显示图例
#训练误差和测试误差
train_err = np.sum(residuals_func(p_lsq[0],x_train,y_train)**2)/len(x_train)
test_err = np.sum(residuals_func(p_lsq[0],x_test,y_test)**2)/len(x_test)
print("训练误差:",train_err)
print("测试误差:",test_err)
return p_lsq,train_err,test_err
# M=0,表示多项式次数为0
p_lsq_0 = fitting3(M=0) #得到的结果为欠拟合
Fitting Parameters: [-0.00254771]
训练误差: 0.5080438309671172
测试误差: 0.5064028740331827
#M = 1
p_lsq_1 = fitting3(M=1)
Fitting Parameters: [-1.9105494 0.95545301]
训练误差: 0.20539699001835074
测试误差: 0.22087267850762224
M=1
# M=3
p_lsq_3 = fitting3(M=3)
Fitting Parameters: [ 23.18756541 -34.78037548 12.00875189 -0.20806997]
训练误差: 0.014295575398024054
测试误差: 0.015509264503001633
M=3
# M=9
p_lsq_9 = fitting3(M=9)
Fitting Parameters: [-3.30005845e+02 1.55625414e+03 -2.95990953e+03 2.89401400e+03 -1.57115336e+03 5.22004345e+02 -1.20865985e+02 3.11880254e+00 6.58323145e+00 -1.51227081e-02]
训练误差: 0.009914936961862256
测试误差: 0.01022899901075852
训练误差就是模型在训练集上的误差平均值,度量了模型对训练集拟合的情况。训练误差大说明对训练集特性学习得不够,训练误差太小说明过度学习了训练集特性,容易发生过拟合。
测试误差是模型在测试集上的误差平均值,度量了模型的泛化能力。在实践中,希望测试误差越小越好。
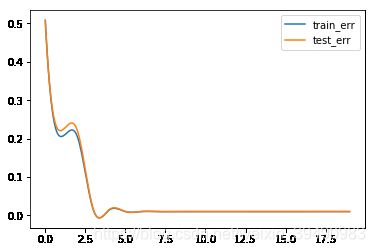
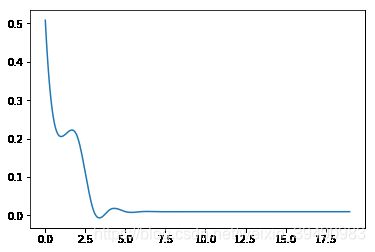
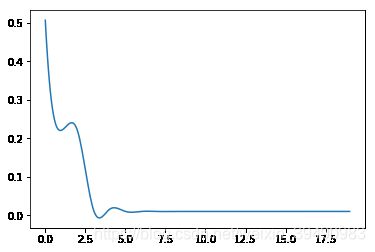
#训练误差,测试误差与模型复杂度,这里我们将多项式次数当作模型复杂度
a = []
b = []
c = []
def err(M=0):
"""
M 为 多项式的最高次数
"""
# 随机初始化多项式参数
p_init = np.random.rand(M+1) #来自均匀分布的长度为M+1的随机数,参数表示指定的形状,均匀分布的属于[0, 1)
# 定义最小二乘法
p_lsq = leastsq(residuals_func, p_init, args=(x_train, y_train)) #来自scipy库的leastsq(),表示最小二乘函数,其中residuals_func为指定的误差函数,p_init为参数初始值,args=(x, y)为误差函数中调用的参数
#leastsq()返回拟合的参数值
#训练误差和测试误差
train_err = np.sum(residuals_func(p_lsq[0],x_train,y_train)**2)/len(x_train)
test_err = np.sum(residuals_func(p_lsq[0],x_test,y_test)**2)/len(x_test)
return train_err,test_err
for i in range(0,20):
train_err = err(M=i)[0]
test_err = err(M=i)[1]
a.append(i)
b.append(train_err)
c.append(test_err)
from scipy.interpolate import spline
a = np.array(a)
new = np.linspace(a.min(),a.max(),300) #300 represents number of points to make between T.min and T.max
b_smooth = spline(a,np.array(b),new)
c_smooth = spline(a,np.array(c),new)
plt.plot(new,b_smooth,label = "train_err")
plt.plot(new,c_smooth,label = "test_err")
plt.legend() #显示图例
plt.plot(new,b_smooth,label = "train_err")
plt.plot(new,c_smooth,label = "test_err")
# M=12
p_lsq_12 = fitting3(M=12)
Fitting Parameters: [ 7.70925250e+02 -2.13948790e+03 1.47955079e+02 5.50179162e+03
-6.90542937e+03 1.62269614e+03 2.85253493e+03 -2.73110094e+03
1.10968608e+03 -2.52646390e+02 1.71394979e+01 5.98163113e+00
-9.12991970e-03]
训练误差: 0.009910459864207754
测试误差: 0.010255984175229287
# M=3
p_lsq_4 = fitting3(M=8)
Fitting Parameters: [ 7.03218670e+01 -1.61073181e+02 3.49309068e+01 1.45531318e+02
-9.15455065e+01 5.06544960e+00 -1.03725482e+01 7.19451627e+00
-2.17477408e-02]
训练误差: 0.009917380173524877
测试误差: 0.010226245482564962
经过简单交叉验证,认为M = 3时,模型效果最好
当模型复杂度增大时,训练误差会减少并趋向于0,测试误差会先减少,达到最小值后又增大。
4、样条插值
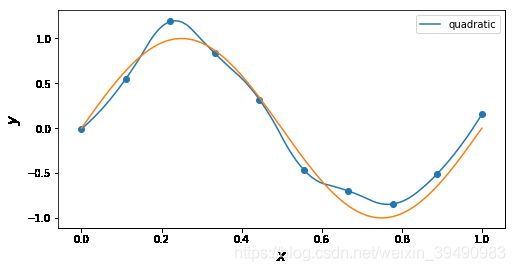
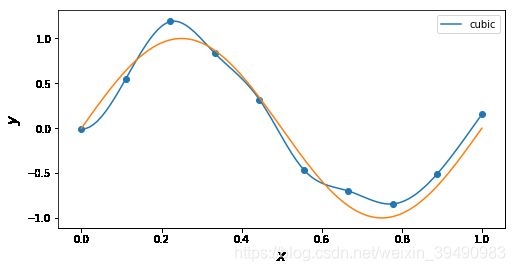
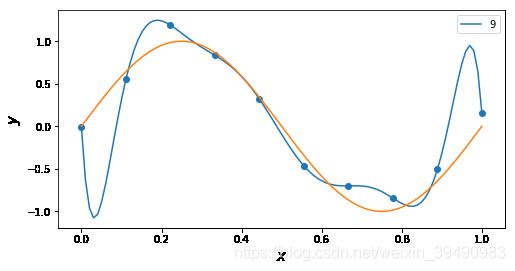
插值与拟合的区别:
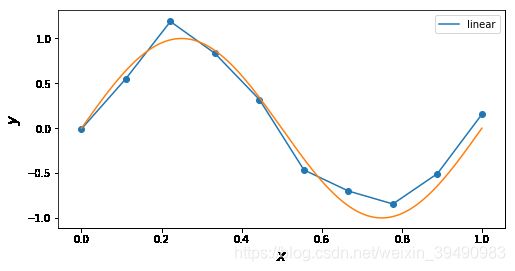
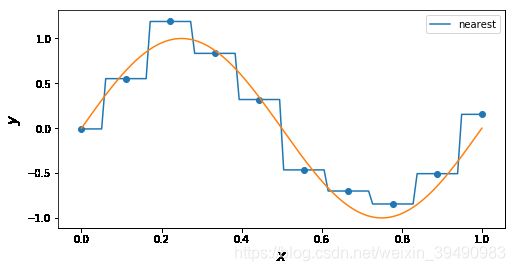
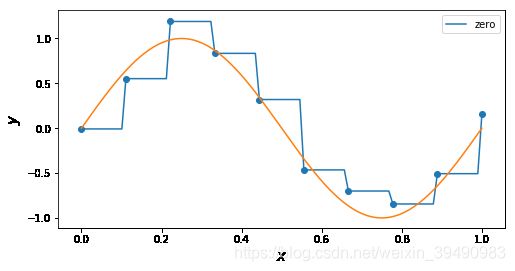
插值曲线要过数据点,拟合曲线不一定要过数据点。
拟合,就是要得到最接近的结果,是要看总体效果。
import scipy.interpolate
import numpy as np, matplotlib.pyplot as plt
from scipy.interpolate import interp1d
#'nearest','zero'为0阶样条插值 ,'
#slinear',‘linear’线性插值 相当于1阶样条插值
#'quadratic'为2阶样条插值
#'cubic'3阶样条插值,更高阶的曲线可以直接使用整数值来指定
x_ = np.linspace(x.min(), x.max(), 100)
fig, ax = plt.subplots(figsize=(8, 4))
ax.scatter(x, y)
for n in ['linear']: # 'linear', 'nearest', 'zero', 'slinear', 'quadratic', 'cubic', 5, 9
f = interp1d(x, y, kind = n)
ax.plot(x_, f(x_), label= n)
ax.legend()
ax.set_ylabel(r"$y$", fontsize=15)
ax.set_xlabel(r"$x$", fontsize=15)
plt.plot(x_points, real_func(x_points), label='real')
plt.show()
x_ = np.linspace(x.min(), x.max(), 100)
fig, ax = plt.subplots(figsize=(8, 4))
ax.scatter(x, y)
for n in [ 'nearest']: # 'linear', 'nearest', 'zero', 'slinear', 'quadratic', 'cubic', 5, 9
f = interp1d(x, y, kind = n)
ax.plot(x_, f(x_), label= n)
ax.legend()
ax.set_ylabel(r"$y$", fontsize=15)
ax.set_xlabel(r"$x$", fontsize=15)
plt.plot(x_points, real_func(x_points), label='real')
plt.show()
x_ = np.linspace(x.min(), x.max(), 100)
fig, ax = plt.subplots(figsize=(8, 4))
ax.scatter(x, y)
for n in [ 'zero']: # 'linear', 'nearest', 'zero', 'slinear', 'quadratic', 'cubic', 5, 9
f = interp1d(x, y, kind = n)
ax.plot(x_, f(x_), label= n)
ax.legend()
ax.set_ylabel(r"$y$", fontsize=15)
ax.set_xlabel(r"$x$", fontsize=15)
plt.plot(x_points, real_func(x_points), label='real')
plt.show()
x_ = np.linspace(x.min(), x.max(), 100)
fig, ax = plt.subplots(figsize=(8, 4))
ax.scatter(x, y)
for n in [ 'slinear']: # 'linear', 'nearest', 'zero', 'slinear', 'quadratic', 'cubic', 5, 9
f = interp1d(x, y, kind = n)
ax.plot(x_, f(x_), label= n)
ax.legend()
ax.set_ylabel(r"$y$", fontsize=15)
ax.set_xlabel(r"$x$", fontsize=15)
plt.plot(x_points, real_func(x_points), label='real')
plt.show()
x_ = np.linspace(x.min(), x.max(), 100)
fig, ax = plt.subplots(figsize=(8, 4))
ax.scatter(x, y)
for n in ['quadratic']: # 'linear', 'nearest', 'zero', 'slinear', 'quadratic', 'cubic', 5, 9
f = interp1d(x, y, kind = n)
ax.plot(x_, f(x_), label= n)
ax.legend()
ax.set_ylabel(r"$y$", fontsize=15)
ax.set_xlabel(r"$x$", fontsize=15)
plt.plot(x_points, real_func(x_points), label='real')
plt.show()
x_ = np.linspace(x.min(), x.max(), 100)
fig, ax = plt.subplots(figsize=(8, 4))
ax.scatter(x, y)
for n in ['cubic']: # 'linear', 'nearest', 'zero', 'slinear', 'quadratic', 'cubic', 5, 9
f = interp1d(x, y, kind = n)
ax.plot(x_, f(x_), label= n)
ax.legend()
ax.set_ylabel(r"$y$", fontsize=15)
ax.set_xlabel(r"$x$", fontsize=15)
plt.plot(x_points, real_func(x_points), label='real')
plt.show()
x_ = np.linspace(x.min(), x.max(), 100)
fig, ax = plt.subplots(figsize=(8, 4))
ax.scatter(x, y)
for n in [5]: # 'linear', 'nearest', 'zero', 'slinear', 'quadratic', 'cubic', 5, 9
f = interp1d(x, y, kind = n)
ax.plot(x_, f(x_), label= n)
ax.legend()
ax.set_ylabel(r"$y$", fontsize=15)
ax.set_xlabel(r"$x$", fontsize=15)
plt.plot(x_points, real_func(x_points), label='real')
plt.show()
x_ = np.linspace(x.min(), x.max(), 100)
fig, ax = plt.subplots(figsize=(8, 4))
ax.scatter(x, y)
for n in [9]: # 'linear', 'nearest', 'zero', 'slinear', 'quadratic', 'cubic', 5, 9
f = interp1d(x, y, kind = n)
ax.plot(x_, f(x_), label= n)
ax.legend()
ax.set_ylabel(r"$y$", fontsize=15)
ax.set_xlabel(r"$x$", fontsize=15)
plt.plot(x_points, real_func(x_points), label='real')
plt.show()
【1】李航.统计学习方法
【2】github https://github.com/wzyonggege/statistical-learning-method/blob/master/LeastSquaresMethod/least_sqaure_method.ipynb