在python中使用SimpleITKeras 进行 mha医学图像加载显示
安装SimpleITK : https://itk.org/Wiki/SimpleITK/GettingStarted#Downloading_the_binaries
打开cmd, 输入pip install SimpleITK
使用SimpleITK
该库显示图片的时候需要第三方图片显示器。这里配置的是ImageJ。
ImageJ 安装(需要安装Java JDK8):
1. https://imagej.nih.gov/ij/download.html下载bundled with64-bit Java 1.8.0_112 版本
2. 解压放到合适的文件夹下,这里我放到C盘下了。
ImageJ配置:
1. 在https://imagej.nih.gov/ij/plugins/nifti.html 上下载nifti_io.jar
2. 找到ImageJ的解压目录,把下载的nifti_io.jar替换掉同名文件。 我的是在C:ij150-win-java8\ImageJ\plugins\Input-Output
3. 重新打开ImageJ,检查File -> Import -> NIfTI-Analyze是否存在。存在的话就说明插件安装成功了。
SimpleITK配置
1. 找到ImageJ.exe存放的目录,拷贝路径,打开cmd
2. 输入 setx SITK_SHOW_COMMAND "C:\blah\blah\ImageJ\ImageJ.exe"
3. 成功的话能看到cmd中输出 SUCCESS: Specified value was saved
4. 在用户环境变量中也能看到ImageJ的运行程序已被加入用户环境变量中
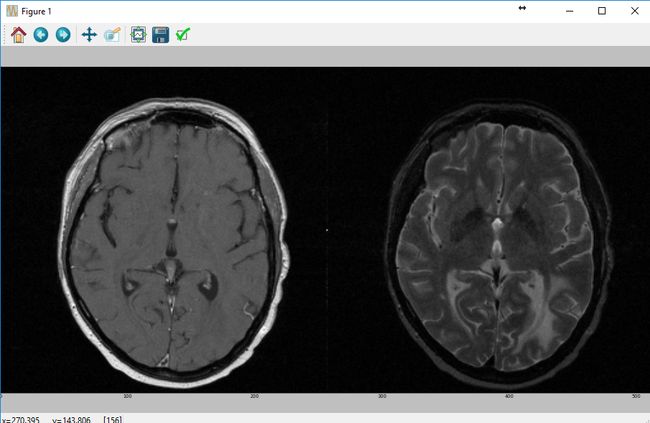
测试配置结果 (mha文件链接:https://bitbucket.org/somada141/pyscience/raw/master/20141016_MultiModalSegmentation/Material/patient_109.zip
测试代码原文链接: https://pyscience.wordpress.com/2014/11/02/multi-modal-image-segmentation-with-python-simpleitk/)
import os
import numpy
import SimpleITK
import matplotlib.pyplot as plt
import numpy
import SimpleITK
import matplotlib.pyplot as plt
def sitk_show(img, title=None, margin=0.0, dpi=40):
nda = SimpleITK.GetArrayFromImage(img)
# spacing = img.GetSpacing()
figsize = (1 + margin) * nda.shape[0] / dpi, (1 + margin) * nda.shape[1] / dpi
# extent = (0, nda.shape[1]*spacing[1], nda.shape[0]*spacing[0], 0)
extent = (0, nda.shape[1], nda.shape[0], 0)
fig = plt.figure(figsize=figsize, dpi=dpi)
ax = fig.add_axes([margin, margin, 1 - 2 * margin, 1 - 2 * margin])
plt.set_cmap("gray")
ax.imshow(nda, extent=extent, interpolation=None)
if title:
plt.title(title)
plt.show()
# Paths to the .mhd files
filenameT1 = "../patient_109/patient_109/mr_T1/patient_109_mr_T1.mhd"
filenameT2 = "../patient_109/patient_109/mr_T2/patient_109_mr_T2.mhd"
# Slice index to visualize with 'sitk_show'
idxSlice = 26
# int label to assign to the segmented gray matter
labelGrayMatter = 1
imgT1Original = SimpleITK.ReadImage(filenameT1)
imgT2Original = SimpleITK.ReadImage(filenameT2)
sitk_show(SimpleITK.Tile(imgT1Original[:, :, idxSlice],
imgT2Original[:, :, idxSlice],
(2, 1, 0)))