python复杂网络分析库networkx
文章目录
-
-
- 1 简介
-
- 安装
- 支持四种图
- 绘制网络图基本流程
- 2 Graph-无向图
-
- 节点
- 边
- 属性
- 有向图和无向图互转
- 3 DiGraph-有向图
- 一些精美的图例子
-
- 环形树状图
- 权重图
- Giant Component
- Random Geometric Graph 随机几何图
- 节点颜色渐变
- 边的颜色渐变
- Atlas
- 画个五角星
- Club
- 画一个多层感知机
- 绘制一个DNN结构图
- 一些图论算法
-
- 最短路径
- 问题
- 一些其他神经网络绘制工具列表
- 参考
-
1 简介
networkx是一个用Python语言开发的图论与复杂网络建模工具,内置了常用的图与复杂网络分析算法,可以方便的进行复杂网络数据分析、仿真建模等工作。
利用networkx可以以标准化和非标准化的数据格式存储网络、生成多种随机网络和经典网络、分析网络结构、建立网络模型、设计新的网络算法、进行网络绘制等。
networkx支持创建简单无向图、有向图和多重图(multigraph);内置许多标准的图论算法,节点可为任意数据;支持任意的边值维度,功能丰富,简单易用。
networkx以图(graph)为基本数据结构。图既可以由程序生成,也可以来自在线数据源,还可以从文件与数据库中读取。
安装
安装的话,跟其他包的安装差不多,用的是anaconda就不用装了。其他就用pip install networkx。
查看版本:
>>> import networkx
>>> networkx.__version__
'1.11'
升级
pip install --upgrade networkx
下面配合使用的一些库,可以选择性安装:
后面可能用到pygraphviz,安装方法如下(亲测有效):
sudo apt-get install graphviz
sudo apt-get install graphviz libgraphviz-dev pkg-config
sudo apt-get install python-pip python-virtualenv
pip install pygraphviz
windows的安装参考这篇博客:
https://blog.csdn.net/fadai1993/article/details/82491657#2____linux_9
安装cv2:
pip install opencv-python #安装非常慢,用下面的方式,从清华源下载
pip3 install -i https://pypi.tuna.tsinghua.edu.cn/simple opencv-python
支持四种图
- Graph:无多重边无向图
- DiGraph:无多重边有向图
- MultiGraph:有多重边无向图
- MultiDiGraph:有多重边有向图
空图对象的创建方式
import networkx as nx
G=nx.Graph()
G=nx.DiGraph()
G=nx.MultiGraph()
G=nx.MultiDiGraph()
G.clear() #清空图
绘制网络图基本流程
- 导入networkx,matplotlib包
- 建立网络
- 绘制网络 nx.draw()
- 建立布局 pos = nx.spring_layout美化作用
#最基本画图程序
import import networkx as nx #导入networkx包
import matplotlib.pyplot as plt
G = nx.random_graphs.barabasi_albert_graph(100,1) #生成一个BA无标度网络G
nx.draw(G) #绘制网络G
plt.savefig("ba.png") #输出方式1: 将图像存为一个png格式的图片文件
plt.show() #输出方式2: 在窗口中显示这幅图像
networkx 提供画图的函数
- draw(G,[pos,ax,hold])
- draw_networkx(G,[pos,with_labels])
- draw_networkx_nodes(G,pos,[nodelist])绘制网络G的节点图
- draw_networkx_edges(G,pos[edgelist])绘制网络G的边图
- draw_networkx_edge_labels(G, pos[, …]) 绘制网络G的边图,边有label
—有layout 布局画图函数的分界线— - draw_circular(G, **kwargs) Draw the graph G with a circular layout.
- draw_random(G, **kwargs) Draw the graph G with a random layout.
- draw_spectral(G, **kwargs)Draw the graph G with a spectral layout.
- draw_spring(G, **kwargs)Draw the graph G with a spring layout.
- draw_shell(G, **kwargs) Draw networkx graph with shell layout.
- draw_graphviz(G[, prog])Draw networkx graph with graphviz layout.
networkx 画图函数里的一些参数
- pos(dictionary, optional): 图像的布局,可选择参数;如果是字典元素,则节点是关键字,位置是对应的值。如果没有指明,则会是spring的布局;也可以使用其他类型的布局,具体可以查阅networkx.layout
- arrows :布尔值,默认True; 对于有向图,如果是True则会画出箭头
- with_labels: 节点是否带标签(默认为True)
- ax:坐标设置,可选择参数;依照设置好的Matplotlib坐标画图
- nodelist:一个列表,默认G.nodes(); 给定节点
- edgelist:一个列表,默认G.edges();给定边
- node_size: 指定节点的尺寸大小(默认是300,单位未知,就是上图中那么大的点)
- node_color: 指定节点的颜色 (默认是红色,可以用字符串简单标识颜色,例如’r’为红色,'b’为绿色等,具体可查看手册),用“数据字典”赋值的时候必须对字典取值(.values())后再赋值
- node_shape: 节点的形状(默认是圆形,用字符串’o’标识,具体可查看手册)
- alpha: 透明度 (默认是1.0,不透明,0为完全透明)
- cmap:Matplotlib的颜色映射,默认None; 用来表示节点对应的强度
- vmin,vmax:浮点数,默认None;节点颜色映射尺度的最大和最小值
- linewidths:[None|标量|一列值];图像边界的线宽
- width: 边的宽度 (默认为1.0)
- edge_color: 边的颜色(默认为黑色)
- edge_cmap:Matplotlib的颜色映射,默认None; 用来表示边对应的强度
- edge_vmin,edge_vmax:浮点数,默认None;边的颜色映射尺度的最大和最小值
- style: 边的样式(默认为实现,可选: solid|dashed|dotted,dashdot)
- labels:字典元素,默认None;文本形式的节点标签
- font_size: 节点标签字体大小 (默认为12)
- font_color: 节点标签字体颜色(默认为黑色)
- node_size:节点大小
- font_weight:字符串,默认’normal’
- font_family:字符串,默认’sans-serif’
布局指定节点排列形式
- circular_layout:节点在一个圆环上均匀分布
- random_layout:节点随机分布
- shell_layout:节点在同心圆上分布
- spring_layout: 用Fruchterman-Reingold算法排列节点,中心放射状分布
- spectral_layout:根据图的拉普拉斯特征向量排列节点
布局也可用pos参数指定,例如,nx.draw(G, pos = spring_layout(G)) 这样指定了networkx上以中心放射状分布.
2 Graph-无向图
如果添加的节点和边是已经存在的,是不会报错的,NetworkX会自动忽略掉已经存在的边和节点的添加。
节点
常用函数
- nodes(G):在图节点上返回一个迭代器
- number_of_nodes(G):返回图中节点的数量
- all_neighbors(graph, node):返回图中节点的所有邻居
- non_neighbors(graph, node):返回图中没有邻居的节点
- common_neighbors(G, u, v):返回图中两个节点的公共邻居
import networkx as nx
import matplotlib.pyplot as plt
G = nx.Graph() # 建立一个空的无向图G
#增加节点
G.add_node('a') # 添加一个节点1
G.add_nodes_from(['b', 'c', 'd', 'e']) # 加点集合
G.add_cycle(['f', 'g', 'h', 'j']) # 加环
H = nx.path_graph(10) # 返回由10个节点的无向图
G.add_nodes_from(H) # 创建一个子图H加入G
G.add_node(H) # 直接将图作为节点
nx.draw(G, with_labels=True,node_color='red')
plt.show()
#访问节点
print('图中所有的节点', G.nodes())
#图中所有的节点 [0, 1, 2, 3, 'a', 'c', 'f', 7, 8, 9, , 'g', 'e', 'h', 'b', 4, 6, 5, 'j', 'd']
print('图中节点的个数', G.number_of_nodes())
#图中节点的个数 20
#删除节点
G.remove_node(1) #删除指定节点
G.remove_nodes_from(['b','c','d','e']) #删除集合中的节点

边
常用函数
- edges(G[, nbunch]):返回与nbunch中的节点相关的边的视图
- number_of_edges(G):返回图中边的数目
- non_edges(graph):返回图中不存在的边
import networkx as nx
import matplotlib.pyplot as plt
#添加边方法1
F = nx.Graph() # 创建无向图
F.add_edge(11,12) #一次添加一条边
#添加边方法2
e=(13,14) #e是一个元组
F.add_edge(*e) #这是python中解包裹的过程
#添加边方法3
F.add_edges_from([(1,2),(1,3)]) #通过添加list来添加多条边
H = nx.path_graph(10) #返回由10个节点的无向图
#通过添加任何ebunch来添加边
F.add_edges_from(H.edges()) #不能写作F.add_edges_from(H)
nx.draw(F, with_labels=True)
plt.show()
#访问边
print('图中所有的边', F.edges())
# 图中所有的边 [(0, 1), (1, 2), (1, 3), (2, 3), (3, 4), (4, 5), (5, 6), (6, 7), (7, 8), (8, 9), (11, 12), (13, 14)]
print('图中边的个数', F.number_of_edges())
# 图中边的个数 12
#删除边
F.remove_edge(1,2)
F.remove_edges_from([(11,12), (13,14)])
nx.draw(F, with_labels=True)
plt.show()

使用邻接迭代器遍历每一条边
import networkx as nx
import matplotlib.pyplot as plt
#快速遍历每一条边,可以使用邻接迭代器实现,对于无向图,每一条边相当于两条有向边
FG = nx.Graph()
FG.add_weighted_edges_from([(1,2,0.125), (1,3,0.75), (2,4,1.2), (3,4,0.275)])
for n, nbrs in FG.adjacency_iter():
for nbr, eattr in nbrs.items():
data = eattr['weight']
print('(%d, %d, %0.3f)' % (n,nbr,data))
# (1, 2, 0.125)
# (1, 3, 0.750)
# (2, 1, 0.125)
# (2, 4, 1.200)
# (3, 1, 0.750)
# (3, 4, 0.275)
# (4, 2, 1.200)
# (4, 3, 0.275)
print('***********************************')
#筛选weight小于0.5的边:
FG = nx.Graph()
FG.add_weighted_edges_from([(1,2,0.125), (1,3,0.75), (2,4,1.2), (3,4,0.275)])
for n, nbrs in FG.adjacency_iter():
for nbr, eattr in nbrs.items():
data = eattr['weight']
if data < 0.5:
print('(%d, %d, %0.3f)' % (n,nbr,data))
# (1, 2, 0.125)
# (2, 1, 0.125)
# (3, 4, 0.275)
# (4, 3, 0.275)
print('***********************************')
#一种方便的访问所有边的方法:
for u,v,d in FG.edges(data = 'weight'):
print((u,v,d))
# (1, 2, 0.125)
# (1, 3, 0.75)
# (2, 4, 1.2)
# (3, 4, 0.275)
属性
- 属性诸如weight,labels,colors,或者任何对象,都可以附加到图、节点或边上。
- 对于每一个图、节点和边都可以在关联的属性字典中保存一个(多个)键-值对。
- 默认情况下这些是一个空的字典,但是可以增加或者是改变这些属性。
图的属性
#图的属性
import networkx as nx
G = nx.Graph(day='Monday') #可以在创建图时分配图的属性
print(G.graph)
G.graph['day'] = 'Friday' #也可以修改已有的属性
print(G.graph)
G.graph['name'] = 'time' #可以随时添加新的属性到图中
print(G.graph)
输出:
{'day': 'Monday'}
{'day': 'Friday'}
{'day': 'Friday', 'name': 'time'}
节点的属性
#节点的属性
import networkx as nx
G = nx.Graph(day='Monday')
G.add_node(1, index='1th') #在添加节点时分配节点属性
# print(G.node(data=True)) #TypeError: 'dict' object is not callable
print(G.node)
#{1: {'index': '1th'}}
G.node[1]['index'] = '0th' #通过G.node[][]来添加或修改属性
print(G.node)
# {1: {'index': '0th'}}
G.add_nodes_from([2,3], index='2/3th') #从集合中添加节点时分配属性
print(G.node)
# {1: {'index': '0th'}, 2: {'index': '2/3th'}, 3: {'index': '2/3th'}}
边的属性
#边的属性
import networkx as nx
G = nx.Graph(day='manday')
G.add_edge(1,2,weight=10) #在添加边时分配属性
print(G.edges(data=True))
#[(1, 2, {'weight': 10})]
G.add_edges_from([(1,3), (4,5)], len=22) #从集合中添加边时分配属性
print(G.edges(data='len'))
# [(1, 2, None), (1, 3, 22), (4, 5, 22)]
G.add_edges_from([(3,4,{'hight':10}),(1,4,{'high':'unknow'})])
print(G.edges(data=True))
# [(1, 2, {'weight': 10}), (1, 3, {'len': 22}), (1, 4, {'high': 'unknow'}), (3, 4, {'hight': 10}), (4, 5, {'len': 22})]
G[1][2]['weight'] = 100000 #通过G[][][]来添加或修改属性
print(G.edges(data=True))
# [(1, 2, {'weight': 100000}), (1, 3, {'len': 22}), (1, 4, {'high': 'unknow'}), (3, 4, {'hight': 10}), (4, 5, {'len': 22})]
有向图和无向图互转
有向图和多重图的基本操作与无向图一致。
无向图与有向图之间可以相互转换,转化方法如下:
#有向图转化成无向图
H=DG.to_undirected()
#或者
H=nx.Graph(DG)
#无向图转化成有向图
F = H.to_directed()
#或者
F = nx.DiGraph(H)
3 DiGraph-有向图
import networkx as nx
import matplotlib.pyplot as plt
G = nx.DiGraph()
G.add_node(1)
G.add_node(2)
G.add_nodes_from([3,4,5,6])
G.add_cycle([1,2,3,4])
G.add_edge(1,3)
G.add_edges_from([(3,5),(3,6),(6,7)])
nx.draw(G,node_color = 'red')
plt.savefig("youxiangtu.png")
plt.show()

from __future__ import division
import matplotlib.pyplot as plt
import networkx as nx
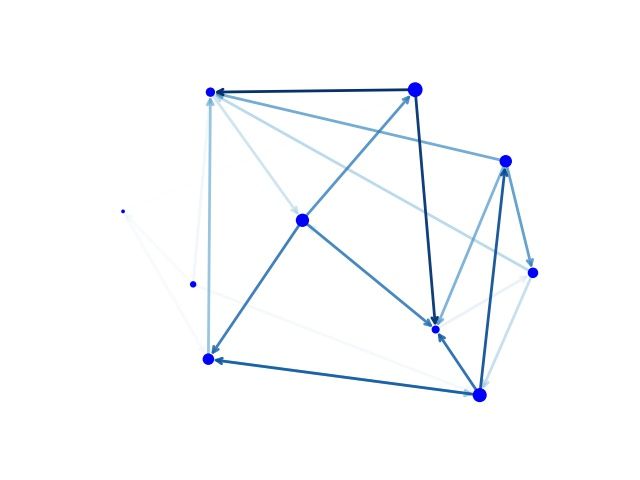
G = nx.generators.directed.random_k_out_graph(10, 3, 0.5)
pos = nx.layout.spring_layout(G)
node_sizes = [3 + 10 * i for i in range(len(G))]
M = G.number_of_edges()
edge_colors = range(2, M + 2)
edge_alphas = [(5 + i) / (M + 4) for i in range(M)]
nodes = nx.draw_networkx_nodes(G, pos, node_size=node_sizes, node_color='blue')
edges = nx.draw_networkx_edges(G, pos, node_size=node_sizes, arrowstyle='->',
arrowsize=10, edge_color=edge_colors,
edge_cmap=plt.cm.Blues, width=2)
# set alpha value for each edge
for i in range(M):
edges[i].set_alpha(edge_alphas[i])
ax = plt.gca()
ax.set_axis_off()
plt.savefig("directed.jpg")
plt.show()
一些精美的图例子
环形树状图
import matplotlib.pyplot as plt
import networkx as nx
try:
import pygraphviz
from networkx.drawing.nx_agraph import graphviz_layout
except ImportError:
try:
import pydot
from networkx.drawing.nx_pydot import graphviz_layout
except ImportError:
raise ImportError("This example needs Graphviz and either "
"PyGraphviz or pydot")
G = nx.balanced_tree(3, 5)
pos = graphviz_layout(G, prog='twopi', args='')
plt.figure(figsize=(8, 8))
nx.draw(G, pos, node_size=20, alpha=0.5, node_color="blue", with_labels=False)
plt.axis('equal')
plt.show()

权重图
import matplotlib.pyplot as plt
import networkx as nx
G = nx.Graph()
G.add_edge('a', 'b', weight=0.6)
G.add_edge('a', 'c', weight=0.2)
G.add_edge('c', 'd', weight=0.1)
G.add_edge('c', 'e', weight=0.7)
G.add_edge('c', 'f', weight=0.9)
G.add_edge('a', 'd', weight=0.3)
elarge = [(u, v) for (u, v, d) in G.edges(data=True) if d['weight'] > 0.5]
esmall = [(u, v) for (u, v, d) in G.edges(data=True) if d['weight'] <= 0.5]
pos = nx.spring_layout(G) # positions for all nodes
# nodes
nx.draw_networkx_nodes(G, pos, node_size=700)
# edges
nx.draw_networkx_edges(G, pos, edgelist=elarge,
width=6)
nx.draw_networkx_edges(G, pos, edgelist=esmall,
width=6, alpha=0.5, edge_color='b', style='dashed')
# labels
nx.draw_networkx_labels(G, pos, font_size=20, font_family='sans-serif')
plt.axis('off')
plt.savefig("weight.jpg")
plt.show()

Giant Component
import math
import matplotlib.pyplot as plt
import networkx as nx
try:
import pygraphviz
from networkx.drawing.nx_agraph import graphviz_layout
layout = graphviz_layout
except ImportError:
try:
import pydot
from networkx.drawing.nx_pydot import graphviz_layout
layout = graphviz_layout
except ImportError:
print("PyGraphviz and pydot not found;\n"
"drawing with spring layout;\n"
"will be slow.")
layout = nx.spring_layout
n = 150 # 150 nodes
# p value at which giant component (of size log(n) nodes) is expected
p_giant = 1.0 / (n - 1)
# p value at which graph is expected to become completely connected
p_conn = math.log(n) / float(n)
# the following range of p values should be close to the threshold
pvals = [0.003, 0.006, 0.008, 0.015]
region = 220 # for pylab 2x2 subplot layout
plt.subplots_adjust(left=0, right=1, bottom=0, top=0.95, wspace=0.01, hspace=0.01)
for p in pvals:
G = nx.binomial_graph(n, p)
pos = layout(G)
region += 1
plt.subplot(region)
plt.title("p = %6.3f" % (p))
nx.draw(G, pos,
with_labels=False,
node_size=10
)
# identify largest connected component
Gcc = sorted(nx.connected_component_subgraphs(G), key=len, reverse=True)
G0 = Gcc[0]
nx.draw_networkx_edges(G0, pos,
with_labels=False,
edge_color='r',
width=6.0
)
# show other connected components
for Gi in Gcc[1:]:
if len(Gi) > 1:
nx.draw_networkx_edges(Gi, pos,
with_labels=False,
edge_color='r',
alpha=0.3,
width=5.0
)
plt.show()

Random Geometric Graph 随机几何图
import matplotlib.pyplot as plt
import networkx as nx
G = nx.random_geometric_graph(200, 0.125)
# position is stored as node attribute data for random_geometric_graph
pos = nx.get_node_attributes(G, 'pos')
# find node near center (0.5,0.5)
dmin = 1
ncenter = 0
for n in pos:
x, y = pos[n]
d = (x - 0.5)**2 + (y - 0.5)**2
if d < dmin:
ncenter = n
dmin = d
# color by path length from node near center
p = dict(nx.single_source_shortest_path_length(G, ncenter))
plt.figure(figsize=(8, 8))
nx.draw_networkx_edges(G, pos, nodelist=[ncenter], alpha=0.4)
nx.draw_networkx_nodes(G, pos, nodelist=list(p.keys()),
node_size=80,
node_color=list(p.values()),
cmap=plt.cm.Reds_r)
plt.xlim(-0.05, 1.05)
plt.ylim(-0.05, 1.05)
#plt.axis('off')
plt.show()
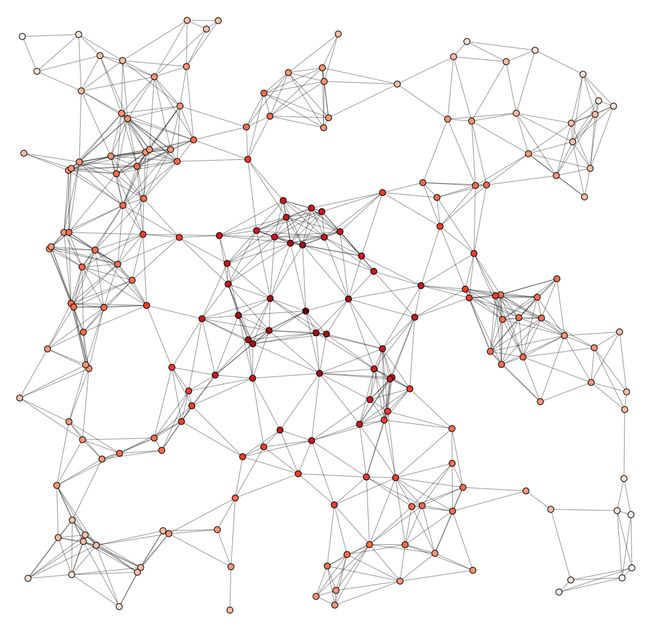
节点颜色渐变
import networkx as nx
import matplotlib.pyplot as plt
G = nx.cycle_graph(24)
pos = nx.spring_layout(G, iterations=200)
nx.draw(G, pos, node_color=range(24), node_size=800, cmap=plt.cm.Blues)
plt.savefig("node.jpg")
plt.show()

边的颜色渐变
import matplotlib.pyplot as plt
import networkx as nx
G = nx.star_graph(20)
pos = nx.spring_layout(G) #布局为中心放射状
colors = range(20)
nx.draw(G, pos, node_color='#A0CBE2', edge_color=colors,
width=4, edge_cmap=plt.cm.Blues, with_labels=False)
plt.show()

Atlas
import random
try:
import pygraphviz
from networkx.drawing.nx_agraph import graphviz_layout
except ImportError:
try:
import pydot
from networkx.drawing.nx_pydot import graphviz_layout
except ImportError:
raise ImportError("This example needs Graphviz and either "
"PyGraphviz or pydot.")
import matplotlib.pyplot as plt
import networkx as nx
from networkx.algorithms.isomorphism.isomorph import graph_could_be_isomorphic as isomorphic
from networkx.generators.atlas import graph_atlas_g
def atlas6():
""" Return the atlas of all connected graphs of 6 nodes or less.
Attempt to check for isomorphisms and remove.
"""
Atlas = graph_atlas_g()[0:208] # 208
# remove isolated nodes, only connected graphs are left
U = nx.Graph() # graph for union of all graphs in atlas
for G in Atlas:
zerodegree = [n for n in G if G.degree(n) == 0]
for n in zerodegree:
G.remove_node(n)
U = nx.disjoint_union(U, G)
# iterator of graphs of all connected components
C = (U.subgraph(c) for c in nx.connected_components(U))
UU = nx.Graph()
# do quick isomorphic-like check, not a true isomorphism checker
nlist = [] # list of nonisomorphic graphs
for G in C:
# check against all nonisomorphic graphs so far
if not iso(G, nlist):
nlist.append(G)
UU = nx.disjoint_union(UU, G) # union the nonisomorphic graphs
return UU
def iso(G1, glist):
"""Quick and dirty nonisomorphism checker used to check isomorphisms."""
for G2 in glist:
if isomorphic(G1, G2):
return True
return False
if __name__ == '__main__':
G = atlas6()
print("graph has %d nodes with %d edges"
% (nx.number_of_nodes(G), nx.number_of_edges(G)))
print(nx.number_connected_components(G), "connected components")
plt.figure(1, figsize=(8, 8))
# layout graphs with positions using graphviz neato
pos = graphviz_layout(G, prog="neato")
# color nodes the same in each connected subgraph
C = (G.subgraph(c) for c in nx.connected_components(G))
for g in C:
c = [random.random()] * nx.number_of_nodes(g) # random color...
nx.draw(g,
pos,
node_size=40,
node_color=c,
vmin=0.0,
vmax=1.0,
with_labels=False
)
plt.show()

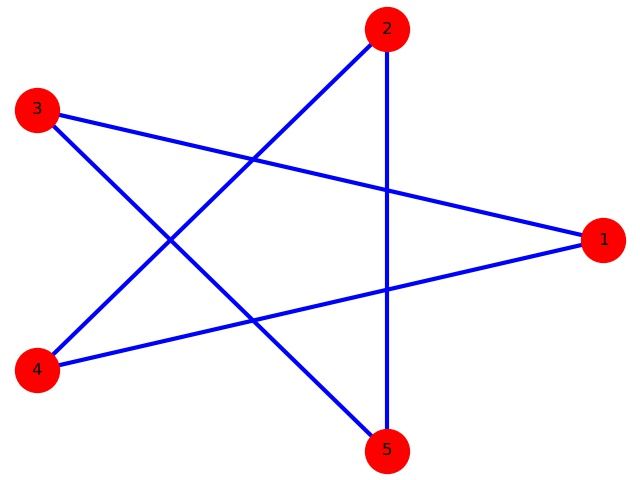
画个五角星
import networkx as nx
import matplotlib.pyplot as plt
#画图!
G=nx.Graph()
G.add_node(1)
G.add_nodes_from([2,3,4,5])
for i in range(5):
for j in range(i):
if (abs(i-j) not in (1,4)):
G.add_edge(i+1, j+1)
nx.draw(G,
with_labels=True, #这个选项让节点有名称
edge_color='b', # b stands for blue!
pos=nx.circular_layout(G), # 这个是选项选择点的排列方式,具体可以用 help(nx.drawing.layout) 查看
# 主要有spring_layout (default), random_layout, circle_layout, shell_layout
# 这里是环形排布,还有随机排列等其他方式
node_color='r', # r = red
node_size=1000, # 节点大小
width=3, # 边的宽度
)
plt.savefig("star.jpg")
plt.show()
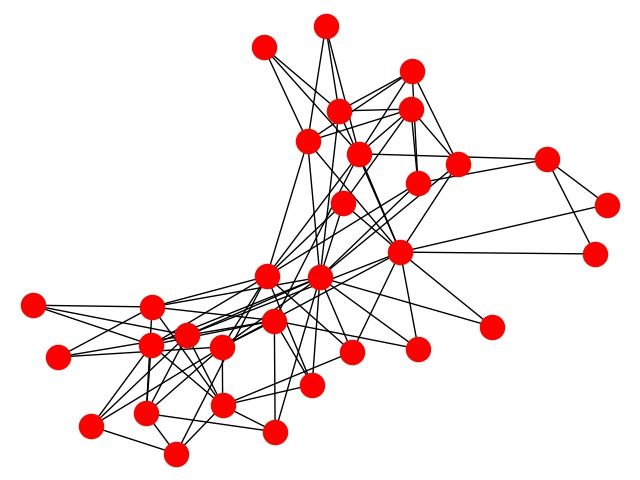
Club
import matplotlib.pyplot as plt
import networkx as nx
import networkx.algorithms.bipartite as bipartite
G = nx.davis_southern_women_graph()
women = G.graph['top']
clubs = G.graph['bottom']
print("Biadjacency matrix")
print(bipartite.biadjacency_matrix(G, women, clubs))
# project bipartite graph onto women nodes
W = bipartite.projected_graph(G, women)
print('')
print("#Friends, Member")
for w in women:
print('%d %s' % (W.degree(w), w))
# project bipartite graph onto women nodes keeping number of co-occurence
# the degree computed is weighted and counts the total number of shared contacts
W = bipartite.weighted_projected_graph(G, women)
print('')
print("#Friend meetings, Member")
for w in women:
print('%d %s' % (W.degree(w, weight='weight'), w))
nx.draw(G,node_color="red")
plt.savefig("club.jpg")
plt.show()
画一个多层感知机
import matplotlib.pyplot as plt
import networkx as nx
left, right, bottom, top, layer_sizes = .1, .9, .1, .9, [4, 7, 7, 2]
# 网络离上下左右的距离
# layter_sizes可以自己调整
import random
G = nx.Graph()
v_spacing = (top - bottom)/float(max(layer_sizes))
h_spacing = (right - left)/float(len(layer_sizes) - 1)
node_count = 0
for i, v in enumerate(layer_sizes):
layer_top = v_spacing*(v-1)/2. + (top + bottom)/2.
for j in range(v):
G.add_node(node_count, pos=(left + i*h_spacing, layer_top - j*v_spacing))
node_count += 1
# 这上面的数字调整我想了好半天,汗
for x, (left_nodes, right_nodes) in enumerate(zip(layer_sizes[:-1], layer_sizes[1:])):
for i in range(left_nodes):
for j in range(right_nodes):
G.add_edge(i+sum(layer_sizes[:x]), j+sum(layer_sizes[:x+1]))
pos=nx.get_node_attributes(G,'pos')
# 把每个节点中的位置pos信息导出来
nx.draw(G, pos,
node_color=range(node_count),
with_labels=True,
node_size=200,
edge_color=[random.random() for i in range(len(G.edges))],
width=3,
cmap=plt.cm.Dark2, # matplotlib的调色板,可以搜搜,很多颜色
edge_cmap=plt.cm.Blues
)
plt.savefig("mlp.jpg")
plt.show()
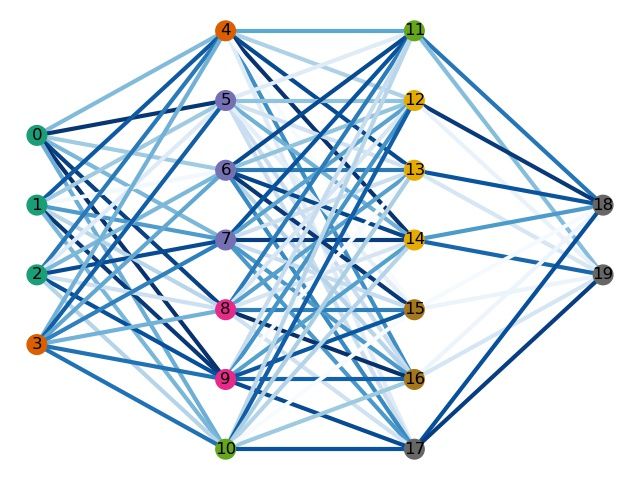
绘制一个DNN结构图
# -*- coding:utf-8 -*-
import networkx as nx
import matplotlib.pyplot as plt
# 创建DAG
G = nx.DiGraph()
# 顶点列表
vertex_list = ['v'+str(i) for i in range(1, 22)]
# 添加顶点
G.add_nodes_from(vertex_list)
# 边列表
edge_list = [
('v1', 'v5'), ('v1', 'v6'), ('v1', 'v7'),('v1', 'v8'),('v1', 'v9'),
('v2', 'v5'), ('v2', 'v6'), ('v2', 'v7'),('v2', 'v8'),('v2', 'v9'),
('v3', 'v5'), ('v3', 'v6'), ('v3', 'v7'),('v3', 'v8'),('v3', 'v9'),
('v4', 'v5'), ('v4', 'v6'), ('v4', 'v7'),('v4', 'v8'),('v4', 'v9'),
('v5','v10'),('v5','v11'),('v5','v12'),('v5','v13'),('v5','v14'),('v5','v15'),
('v6','v10'),('v6','v11'),('v6','v12'),('v6','v13'),('v6','v14'),('v6','v15'),
('v7','v10'),('v7','v11'),('v7','v12'),('v7','v13'),('v7','v14'),('v7','v15'),
('v8','v10'),('v8','v11'),('v8','v12'),('v8','v13'),('v8','v14'),('v8','v15'),
('v9','v10'),('v9','v11'),('v9','v12'),('v9','v13'),('v9','v14'),('v9','v15'),
('v10','v16'),('v10','v17'),('v10','v18'),
('v11','v16'),('v11','v17'),('v11','v18'),
('v12','v16'),('v12','v17'),('v12','v18'),
('v13','v16'),('v13','v17'),('v13','v18'),
('v14','v16'),('v14','v17'),('v14','v18'),
('v15','v16'),('v15','v17'),('v15','v18'),
('v16','v19'),
('v17','v20'),
('v18','v21')
]
# 通过列表形式来添加边
G.add_edges_from(edge_list)
# 绘制DAG图
plt.title('DNN for iris') #图片标题
nx.draw(
G,
node_color = 'red', # 顶点颜色
edge_color = 'black', # 边的颜色
with_labels = True, # 显示顶点标签
font_size =10, # 文字大小
node_size =300 # 顶点大小
)
# 显示图片
plt.show()
可以看到,在代码中已经设置好了这22个神经元以及它们之间的连接情况,但绘制出来的结构如却是这样的:

这显然不是想要的结果,因为各神经的连接情况不明朗,而且很多神经都挤在了一起,看不清楚。之所以出现这种情况,是因为没有给神经元设置坐标,导致每个神经元都是随机放置的。
接下来,引入坐标机制,即设置好每个神经元节点的坐标,使得它们的位置能够按照事先设置好的来放置,其Python代码如下:
# -*- coding:utf-8 -*-
import networkx as nx
import matplotlib.pyplot as plt
# 创建DAG
G = nx.DiGraph()
# 顶点列表
vertex_list = ['v'+str(i) for i in range(1, 22)]
# 添加顶点
G.add_nodes_from(vertex_list)
# 边列表
edge_list = [
('v1', 'v5'), ('v1', 'v6'), ('v1', 'v7'),('v1', 'v8'),('v1', 'v9'),
('v2', 'v5'), ('v2', 'v6'), ('v2', 'v7'),('v2', 'v8'),('v2', 'v9'),
('v3', 'v5'), ('v3', 'v6'), ('v3', 'v7'),('v3', 'v8'),('v3', 'v9'),
('v4', 'v5'), ('v4', 'v6'), ('v4', 'v7'),('v4', 'v8'),('v4', 'v9'),
('v5','v10'),('v5','v11'),('v5','v12'),('v5','v13'),('v5','v14'),('v5','v15'),
('v6','v10'),('v6','v11'),('v6','v12'),('v6','v13'),('v6','v14'),('v6','v15'),
('v7','v10'),('v7','v11'),('v7','v12'),('v7','v13'),('v7','v14'),('v7','v15'),
('v8','v10'),('v8','v11'),('v8','v12'),('v8','v13'),('v8','v14'),('v8','v15'),
('v9','v10'),('v9','v11'),('v9','v12'),('v9','v13'),('v9','v14'),('v9','v15'),
('v10','v16'),('v10','v17'),('v10','v18'),
('v11','v16'),('v11','v17'),('v11','v18'),
('v12','v16'),('v12','v17'),('v12','v18'),
('v13','v16'),('v13','v17'),('v13','v18'),
('v14','v16'),('v14','v17'),('v14','v18'),
('v15','v16'),('v15','v17'),('v15','v18'),
('v16','v19'),
('v17','v20'),
('v18','v21')
]
# 通过列表形式来添加边
G.add_edges_from(edge_list)
# 指定绘制DAG图时每个顶点的位置
pos = {
'v1':(-2,1.5),
'v2':(-2,0.5),
'v3':(-2,-0.5),
'v4':(-2,-1.5),
'v5':(-1,2),
'v6': (-1,1),
'v7':(-1,0),
'v8':(-1,-1),
'v9':(-1,-2),
'v10':(0,2.5),
'v11':(0,1.5),
'v12':(0,0.5),
'v13':(0,-0.5),
'v14':(0,-1.5),
'v15':(0,-2.5),
'v16':(1,1),
'v17':(1,0),
'v18':(1,-1),
'v19':(2,1),
'v20':(2,0),
'v21':(2,-1)
}
# 绘制DAG图
plt.title('DNN for iris') #图片标题
plt.xlim(-2.2, 2.2) #设置X轴坐标范围
plt.ylim(-3, 3) #设置Y轴坐标范围
nx.draw(
G,
pos = pos, # 点的位置
node_color = 'red', # 顶点颜色
edge_color = 'black', # 边的颜色
with_labels = True, # 显示顶点标签
font_size =10, # 文字大小
node_size =300 # 顶点大小
)
# 显示图片
plt.show()
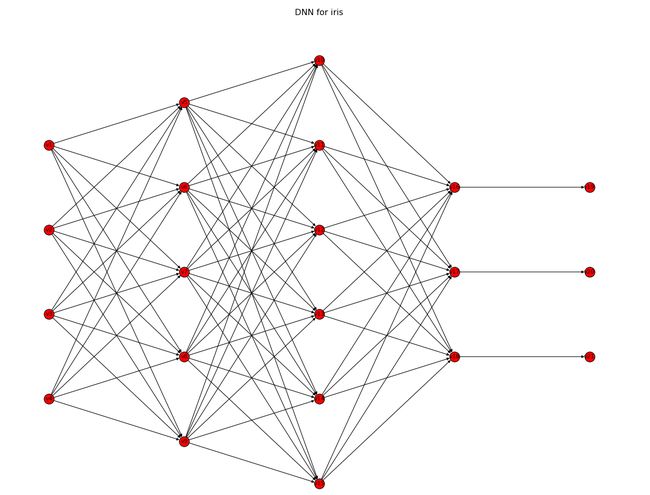
可以看到,在代码中,通过pos字典已经规定好了每个神经元节点的位置。
接下来,需要对这个框架图进行更为细致地修改,需要修改的地方为:
- 去掉神经元节点的标签;
- 添加模型层的文字注释(比如Input layer)
其中,第二步的文字注释,我们借助opencv来完成。完整的Python代码如下:
# -*- coding:utf-8 -*-
import cv2
import networkx as nx
import matplotlib.pyplot as plt
# 创建DAG
G = nx.DiGraph()
# 顶点列表
vertex_list = ['v'+str(i) for i in range(1, 22)]
# 添加顶点
G.add_nodes_from(vertex_list)
# 边列表
edge_list = [
('v1', 'v5'), ('v1', 'v6'), ('v1', 'v7'),('v1', 'v8'),('v1', 'v9'),
('v2', 'v5'), ('v2', 'v6'), ('v2', 'v7'),('v2', 'v8'),('v2', 'v9'),
('v3', 'v5'), ('v3', 'v6'), ('v3', 'v7'),('v3', 'v8'),('v3', 'v9'),
('v4', 'v5'), ('v4', 'v6'), ('v4', 'v7'),('v4', 'v8'),('v4', 'v9'),
('v5','v10'),('v5','v11'),('v5','v12'),('v5','v13'),('v5','v14'),('v5','v15'),
('v6','v10'),('v6','v11'),('v6','v12'),('v6','v13'),('v6','v14'),('v6','v15'),
('v7','v10'),('v7','v11'),('v7','v12'),('v7','v13'),('v7','v14'),('v7','v15'),
('v8','v10'),('v8','v11'),('v8','v12'),('v8','v13'),('v8','v14'),('v8','v15'),
('v9','v10'),('v9','v11'),('v9','v12'),('v9','v13'),('v9','v14'),('v9','v15'),
('v10','v16'),('v10','v17'),('v10','v18'),
('v11','v16'),('v11','v17'),('v11','v18'),
('v12','v16'),('v12','v17'),('v12','v18'),
('v13','v16'),('v13','v17'),('v13','v18'),
('v14','v16'),('v14','v17'),('v14','v18'),
('v15','v16'),('v15','v17'),('v15','v18'),
('v16','v19'),
('v17','v20'),
('v18','v21')
]
# 通过列表形式来添加边
G.add_edges_from(edge_list)
# 指定绘制DAG图时每个顶点的位置
pos = {
'v1':(-2,1.5),
'v2':(-2,0.5),
'v3':(-2,-0.5),
'v4':(-2,-1.5),
'v5':(-1,2),
'v6': (-1,1),
'v7':(-1,0),
'v8':(-1,-1),
'v9':(-1,-2),
'v10':(0,2.5),
'v11':(0,1.5),
'v12':(0,0.5),
'v13':(0,-0.5),
'v14':(0,-1.5),
'v15':(0,-2.5),
'v16':(1,1),
'v17':(1,0),
'v18':(1,-1),
'v19':(2,1),
'v20':(2,0),
'v21':(2,-1)
}
# 绘制DAG图
plt.title('DNN for iris') #图片标题
plt.xlim(-2.2, 2.2) #设置X轴坐标范围
plt.ylim(-3, 3) #设置Y轴坐标范围
nx.draw(
G,
pos = pos, # 点的位置
node_color = 'red', # 顶点颜色
edge_color = 'black', # 边的颜色
font_size =10, # 文字大小
node_size =300 # 顶点大小
)
# 保存图片,图片大小为640*480
plt.savefig('DNN_sketch.png')
# 利用opencv模块对DNN框架添加文字注释
# 读取图片
imagepath = 'DNN_sketch.png'
image = cv2.imread(imagepath, 1)
# 输入层
cv2.rectangle(image, (85, 130), (120, 360), (255,0,0), 2)
cv2.putText(image, "Input Layer", (15, 390), 1, 1.5, (0, 255, 0), 2, 1)
# 隐藏层
cv2.rectangle(image, (190, 70), (360, 420), (255,0,0), 2)
cv2.putText(image, "Hidden Layer", (210, 450), 1, 1.5, (0, 255, 0), 2, 1)
# 输出层
cv2.rectangle(image, (420, 150), (460, 330), (255,0,0), 2)
cv2.putText(image, "Output Layer", (380, 360), 1, 1.5, (0, 255, 0), 2, 1)
# sofrmax层
cv2.rectangle(image, (530, 150), (570, 330), (255,0,0), 2)
cv2.putText(image, "Softmax Func", (450, 130), 1, 1.5, (0, 0, 255), 2, 1)
# 保存修改后的图片
cv2.imwrite('DNN.png', image)
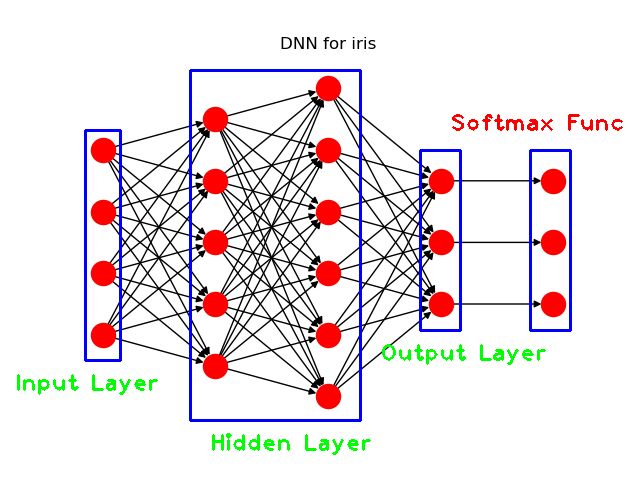
一些图论算法
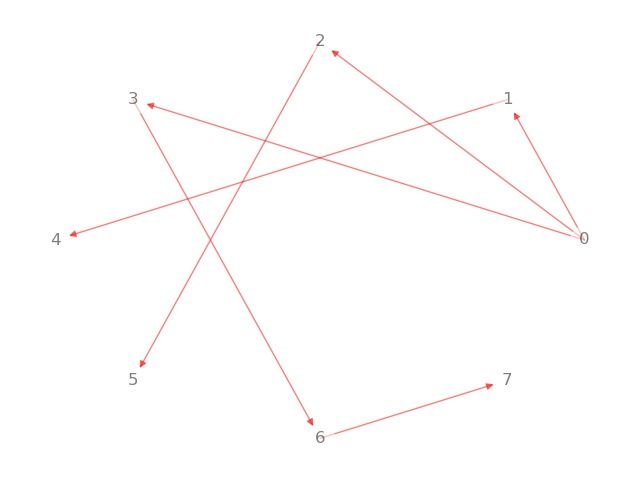
最短路径
函数调用:
dijkstra_path(G, source, target, weight=‘weight’) ————求最短路径
dijkstra_path_length(G, source, target, weight=‘weight’) ————求最短距离
import networkx as nx
import pylab
import numpy as np
#自定义网络
row=np.array([0,0,0,1,2,3,6])
col=np.array([1,2,3,4,5,6,7])
value=np.array([1,2,1,8,1,3,5])
print('生成一个空的有向图')
G=nx.DiGraph()
print('为这个网络添加节点...')
for i in range(0,np.size(col)+1):
G.add_node(i)
print('在网络中添加带权中的边...')
for i in range(np.size(row)):
G.add_weighted_edges_from([(row[i],col[i],value[i])])
print('给网路设置布局...')
pos=nx.shell_layout(G)
print('画出网络图像:')
nx.draw(G,pos,with_labels=True, node_color='white', edge_color='red', node_size=400, alpha=0.5 )
pylab.title('Self_Define Net',fontsize=15)
pylab.show()
'''
Shortest Path with dijkstra_path
'''
print('dijkstra方法寻找最短路径:')
path=nx.dijkstra_path(G, source=0, target=7)
print('节点0到7的路径:', path)
print('dijkstra方法寻找最短距离:')
distance=nx.dijkstra_path_length(G, source=0, target=7)
print('节点0到7的距离为:', distance)
输出:
生成一个空的有向图
为这个网络添加节点...
在网络中添加带权中的边...
给网路设置布局...
画出网络图像:
dijkstra方法寻找最短路径:
节点0到7的路径: [0, 3, 6, 7]
dijkstra方法寻找最短距离:
节点0到7的距离为: 9
问题
本人在pycharm中运行下列程序:
import networkx as nx
import matplotlib.pyplot as plt
G = nx.Graph() # 建立一个空的无向图G
G.add_node('a') # 添加一个节点1
G.add_nodes_from(['b', 'c', 'd', 'e']) # 加点集合
G.add_cycle(['f', 'g', 'h', 'j']) # 加环
H = nx.path_graph(10) # 返回由10个节点挨个连接的无向图,所以有9条边
G.add_nodes_from(H) # 创建一个子图H加入G
G.add_node(H) # 直接将图作为节点
nx.draw(G, with_labels=True)
plt.show()
发现在Pycharm下使用matploylib库绘制3D图的时候,在最后需要显示图像的时候,每当输入plt.show() 都会报错
plt.show()
/yyl/Python/3.6/lib/python/site-packages/matplotlib/figure.py:1743: UserWarning: This figure includes Axes that are not compatible with tight_layout, so its results might be incorrect.
warnings.warn("This figure includes Axes that are not "
...
ValueError: max() arg is an empty sequence
网上的解决方案:
File -> Setting -> Tools -> Python Scientific中去掉对Show plots in tool window的勾选就好了
一些其他神经网络绘制工具列表
| 名称 | 描述 | 推荐指数 |
|---|---|---|
| Python+Graphviz | graphviz的python版本(亲测) | **** |
| PlotNeuralNet | 第一步生成tex文件,然后调用LaTeX命令行生成图形(亲测) | **** |
| NetworkX | 这是个专门的复杂网络图的Python包 | **** |
| Matplotlib’s Viznet | 利用Matplotlib的Viznet | **** |
| LaTeX tikz | 绘制网络结点图的tikz库 | *** |
| Graphviz | 专业绘图软件,dot描述语言 | *** |
| Inkscape | 属于绘图软件 | *** |
| Omnigraffle | 由The Omni Group制作的一款绘图软件 | *** |
| netron | 支持ONNX (.onnx, .pb, .pbtxt), Keras (.h5, .keras), CoreML (.mlmodel), Caffe2 (predict_net.pb, predict_net.pbtxt), MXNet (.model, -symbol.json) and TensorFlow Lite (.tflite),在前面链接处下载文件,在这里演示 | *** |
| TensorBoard | 配合Tensorflow一起使用的 | *** |
| Keras | 自带plot方法,例子,也需要安装graphviz,pydot等 | *** |
| Netscope for Caffe | 只支持Caffe格式,例子 | *** |
| draw_convnet | 这个其实是利用Matplotlib的绘图功能写了一个Python脚本 | ** |
| dnngraph | 仅使用于Caffe框架 | ** |
| ConvNetDraw | 灵活性差,没有更新了 | ** |
上面都是一些这个网络库使用的一点总结,更多内容可以参考下面的官方链接。
参考
- 官方教程:https://networkx.github.io/documentation/stable/_downloads/networkx_reference.pdf
- 官方网站:https://networkx.github.io/documentation/latest/index.html
- 官方githu博客:http://networkx.github.io/
- 用Python的networkx绘制精美网络图
- networkx整理
- Networkx使用指南
- 论文中绘制神经网络工具汇总
- networkx + Cytoscape 构建及可视化网络图
- 用python+graphviz/networkx画目录结构树状图