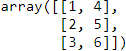cp10_回归预测连续目标变量_boston_Residual_plot_mlxtend_sns_pd_covariance_correlation_RANSAC_R2_Ridge_C_F_A_K_树
Predicting Continuous Target Variables with Regression Analysis
Throughout the previous https://blog.csdn.net/Linli522362242/article/details/110740856(cp9_Embedding a Model into a Web Application_pickle_sqlite3_Flask_wtforms_pythonanywhere_pickle_serialization), you learned a lot about the main concepts behind supervised learning and trained many different models for classification tasks to predict group memberships or categorical variables. We will dive into another subcategory of supervised learning: regression analysis.
Regression models are used to predict target variables on a continuous scale, which makes them attractive for addressing many questions in science. They also have applications in industry, such as understanding relationships between variables, evaluating trends, or making forecasts. One example is predicting the sales of a company in future months.
In this chapter, we will discuss the main concepts of regression models and cover the following topics:
• Exploring and visualizing datasets
• Looking at different approaches to implement linear regression models
• Training regression models that are robust to outliers
• Evaluating regression models and diagnosing common problems
• Fitting regression models to nonlinear data
Introducing linear regression
The goal of linear regression is to model the relationship between one or multiple features and a continuous target variable. In contrast to classification—a different subcategory of supervised learning—regression analysis aims to predict outputs on a continuous scale rather than categorical class labels.
In the following subsections, you will be introduced to the most basic type of linear regression, simple linear regression, and understand how to relate it to the more general, multivariate case (linear regression with multiple features).
Simple linear regression
The goal of simple (univariate) linear regression is to model the relationship between a single feature (explanatory variable, x) and a continuous-valued target (response variable, y). The equation of a linear model with one explanatory variable is defined as follows:![]()
Here, the weight, ![]() , represents the y axis intercept and
, represents the y axis intercept and ![]() is the weight coefficient of the explanatory variable. Our goal is to learn the weights of the linear equation to describe the relationship between the explanatory variable and the target variable, which can then be used to predict the responses of new explanatory variables that were not part of the training dataset.
is the weight coefficient of the explanatory variable. Our goal is to learn the weights of the linear equation to describe the relationship between the explanatory variable and the target variable, which can then be used to predict the responses of new explanatory variables that were not part of the training dataset.
Based on the linear equation that we defined previously, linear regression can be understood as finding the best-fitting straight line through the training examples, as shown in the following figure:![]() https://blog.csdn.net/Linli522362242/article/details/104005906
https://blog.csdn.net/Linli522362242/article/details/104005906
This best-fitting line is also called the regression line, and the vertical lines from the regression line to the training examples are the so-called offsets or residuals—the errors of our prediction.
Multiple linear regression
The previous section introduced simple linear regression, a special case of linear regression with one explanatory variable. Of course, we can also generalize the linear regression model to multiple explanatory variables; this process is called multiple linear regression:
Here, ![]() is the y axis intercept with
is the y axis intercept with ![]() = 1.
= 1.
The following figure shows how the two-dimensional, fitted hyperplane of a multiple linear regression model with two features could look:

As you can see, visualizations of multiple linear regression hyperplanes in a three-dimensional scatterplot are already challenging to interpret when looking at static figures. Since we have no good means of visualizing hyperplanes with two dimensions in a scatterplot (multiple linear regression models fit to datasets with three or more features), the examples and visualizations in this chapter will mainly focus on the univariate case, using simple linear regression. However, simple and multiple linear regression are based on the same concepts and the same evaluation
techniques; the code implementations that we will discuss in this chapter are also compatible with both types of regression model.
Exploring the Housing dataset
Before we implement the first linear regression model, we will discuss a new dataset, the Housing dataset, which contains information about houses in the suburbs of Boston collected by D. Harrison and D.L. Rubinfeld in 1978. The Housing dataset has been made freely available and is included in the code bundle of this book. The dataset has recently been removed from the UCI Machine Learning Repository but is available at (from tensorflow.keras.datasets import boston_housing 10_Introduction to Artificial Neural Networks w Keras1_HuberLoss_astype_dtype_DNN_MLP_G.gv.pdf_EMA https://blog.csdn.net/Linli522362242/article/details/106433059 OR https://blog.csdn.net/Linli522362242/article/details/106755312) or scikit-learn (https://github.com/scikit-learn/scikit-learn/blob/master/sklearn/datasets/data/boston_house_prices.csv). As with each new dataset, it is always helpful to explore the data through a simple visualization, to get a better feeling of what we are working with.
Loading the Housing dataset into a data frame
In this section, we will load the Housing dataset using the pandas read_csv function, which is fast and versatile and a recommended tool for working with tabular data stored in a plaintext format.
The features of the 506 examples in the Housing dataset have been taken from the original source that was previously shared on https://archive.ics.uci.edu/ml/datasets/Housing and summarized here:
- • CRIM: Per capita crime rate by town 城镇人均犯罪率
- • ZN: Proportion of residential land zoned for lots over 25,000 sq. ft. 占地远超过25,000平方英尺的住宅用地比例
- • INDUS: Proportion of non-retail business acres['eɪkəs]英亩 per town 每个镇非零售业务用地比例
- • CHAS: Charles River dummy variable (= 1 if tract bounds束缚 river and 0 otherwise如果房屋位于河边则值为1,否则为0)
If a qualitative predictor定性预测因子 (also known as a factor) only has two levels, or possible values, then incorporating it into a regression model is very simple. We level simply create an indicator or dummy variable that takes on two possible numerical values. For example, based on the gender variable, we can create variable a new variable that takes the form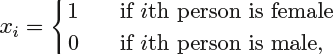
- • NOX: Nitric oxide concentration (parts per 10 million) 一氧化氮浓度(每千万分之一)
- • RM: Average number of rooms per dwelling[ˈdwelɪŋ]住宅
- • AGE: Proportion of owner-occupied units自住单元 built prior to 1940
- • DIS: Weighted distances to five Boston employment centers
- • RAD: Index of accessibility to radial highways 径向公路通达性指数
- • TAX: Full-value property tax rate per $10,000 每10,000美元的全值财产税率
- • PTRATIO: Pupil-teacher ratio by town 各镇师生比例
- • B: 1000*
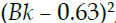 , where Bk is the proportion of [people of African American descent血统] by town
, where Bk is the proportion of [people of African American descent血统] by town - • LSTAT: Percentage of lower status地位 of the population
- • MEDV: Median value of owner-occupied homes in $1000s 自住房的中位数价值(以1000美元计)
For the rest of this chapter, we will regard the house prices (MEDV) as our target variable—the variable that we want to predict using one or more of the 13 explanatory variables. Before we explore this dataset further, let's load it into a pandas DataFrame:
https://raw.githubusercontent.com/scikit-learn/scikit-learn/master/sklearn/datasets/data/boston_house_prices.csv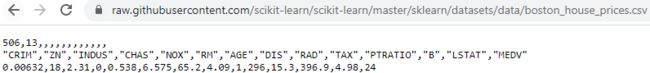
import pandas as pd
df = pd.read_csv('https://raw.githubusercontent.com/scikit-learn/scikit-learn/master/sklearn/datasets/data/boston_house_prices.csv',
header=1,
sep=',')
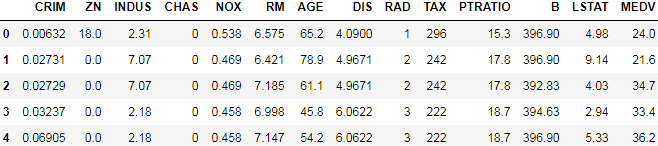
df.head()To confirm that the dataset was loaded successfully, we can display the first five lines of the dataset, as shown in the following figure:
df.columnsIndex(['CRIM', 'ZN', 'INDUS', 'CHAS', 'NOX', 'RM', 'AGE', 'DIS', 'RAD', 'TAX', 'PTRATIO', 'B', 'LSTAT', 'MEDV'],
dtype='object')
Visualizing the important characteristics of a dataset
Exploratory data analysis (EDA) is an important and recommended first step prior to the training of a machine learning model. In the rest of this section, we will use some simple yet useful techniques from the graphical EDA toolbox that may help us to visually detect the presence of outliers, the distribution of the data, and the relationships between features.
First, we will create a scatterplot matrix that allows us to visualize the pair-wise correlations between the different features in this dataset in one place. To plot the scatterplot matrix, we will use the scatterplot matrix function from the MLxtend library (http://rasbt.github.io/mlxtend/), which is a Python library that contains various convenience functions for machine learning and data science
applications in Python.
You can install the mlxtend package via conda install mlxtend or pip install mlxtend(https://blog.csdn.net/Linli522362242/article/details/108037567). After the installation is complete, you can import the package and create the scatterplot matrix as follows:
mlxtend.plotting VS seaborn VS pandas.plotting
import matplotlib.pyplot as plt
from mlxtend.plotting import scatterplotmatrix
cols = ['LSTAT', # Percentage of lower status of the population
'INDUS', # Proportion of non-retail business acres per town
'NOX', # Nitric oxide concentration (parts per 10 million)
'RM', # Average number of rooms per dwelling
'MEDV'] # Median value of owner-occupied homes in $1000s
scatterplotmatrix( df[cols].values, figsize=(10,8), names=cols, alpha=0.5 )
plt.tight_layout()
plt.show()As you can see in the following figure, the scatterplot matrix provides us with a useful graphical summary of the relationships in a dataset:

mlxtend.plotting VS seaborn VS pandas.plotting
Using this scatterplot matrix, we can now quickly eyeball how the data is distributed and whether it contains outliers. For example, we can see that there is a linear relationship between RM and the housing prices MEDV (the fifth column of the fourth row)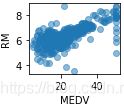 . Furthermore, we can see in the histogram (the lower right subplot in the scatter plot matrix) that the MEDV variable seems to be normally distributed but contains several outliers
. Furthermore, we can see in the histogram (the lower right subplot in the scatter plot matrix) that the MEDV variable seems to be normally distributed but contains several outliers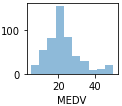 .
.
####################################
Normality assumption of linear regression
Note that in contrast to common belief, training a linear regression model does not require that the explanatory or target variables are normally distributed. The normality assumption is only a requirement for certain statistics and hypothesis tests that are beyond the scope of this book (for more information on this topic, please refer to Introduction to Linear Regression Analysis, Montgomery, Douglas C. Montgomery, Elizabeth A. Peck, and G.Geoffrey Vining, Wiley, 2012, pages: 318-319).
####################################
Figure 2-14 shows various plots along with the correlation coefficient between their horizontal and vertical axes https://blog.csdn.net/Linli522362242/article/details/103387527
import matplotlib.pyplot as plt
import seaborn as sns
cols = ['LSTAT', # Percentage of lower status of the population
'INDUS', # Proportion of non-retail business acres per town
'NOX', # Nitric oxide concentration (parts per 10 million)
'RM', # Average number of rooms per dwelling
'MEDV'] # Median value of owner-occupied homes in $1000s
sns.set( style='whitegrid', context='notebook')
sns.pairplot( df[cols], height=2.0)
plt.show()Due to space constraints and in the interest of readability, we have only plotted five columns from the dataset: LSTAT, INDUS, NOX, RM, and MEDV. However, you are encouraged to create a scatterplot matrix of the whole DataFrame to explore the dataset further by choosing different column names in the previous scatterplotmatrix function call, or including all variables in the scatterplot matrix by omitting the column selector.

######################################################################
Importing the seaborn library modifies the default aesthetics of matplotlib for the current Python session. If you do not want to use seaborn's style settings, you can reset the matplotlib settings
by executing the following command:
sns.reset_orig()######################################################################
mlxtend.plotting VS seaborn VS pandas.plotting
from pandas.plotting import scatter_matrix
import matplotlib.pyplot as plt
cols = ['LSTAT', # Percentage of lower status of the population
'INDUS', # Proportion of non-retail business acres per town
'NOX', # Nitric oxide concentration (parts per 10 million)
'RM', # Average number of rooms per dwelling
'MEDV'] # Median value of owner-occupied homes in $1000s
scatter_matrix( df[cols], figsize=(12,10) )
plt.show()Looking at relationships using a correlation matrix相关系数矩阵
In the previous section, we visualized the data distributions of the Housing dataset variables in the form of histograms and scatterplots. Next, we will create a correlation matrix to quantify量化 and summarize linear relationships between variables https://blog.csdn.net/Linli522362242/article/details/103387527. A correlation matrix is closely related to the covariance matrix that we covered in the section Unsupervised dimensionality reduction via principal component analysis in cp5_Compressing Data via Dimensionality Reduction_feature extraction_PCA_LDA_convergence_kernel PCA https://blog.csdn.net/Linli522362242/article/details/105196037. We can interpret the correlation matrix as being a rescaled version of the covariance matrix. In fact, the correlation matrix is identical to a covariance matrix computed from standardized features.
###########################################################
https://blog.csdn.net/Linli522362242/article/details/105196037
standardize the features(the PCA directions are highly sensitive to data scaling, and we need to standardize the features prior to PCA if the features were measured on different scales and we want to assign equal importance to all features.):
# standardize the features
from sklearn.preprocessing import StandardScaler
sc = StandardScaler()
X_train_std = sc.fit_transform(X_train)
X_test_std = sc.transform(X_test) second step: constructing the covariance matrix. The symmetric d × d -dimensional covariance matrix, where d is the number of dimensions in the dataset, stores the pairwise成对地 covariances between the different features. For example, the covariance协方差 between two features ![]() and
and ![]() on the population level can be calculated via the following equation:
on the population level can be calculated via the following equation:![]()
Here, ![]() and
and ![]() are the sample means of feature j and k , respectively. Note that the sample means are zero if we standardize the dataset. A positive covariance between two features indicates that the features increase or decrease together, whereas a negative covariance indicates that the features vary in opposite directions. For example, a covariance matrix of three features can then be written as (note that
are the sample means of feature j and k , respectively. Note that the sample means are zero if we standardize the dataset. A positive covariance between two features indicates that the features increase or decrease together, whereas a negative covariance indicates that the features vary in opposite directions. For example, a covariance matrix of three features can then be written as (note that ![]() stands for the Greek uppercase letter sigma, which is not to be confused with the sum symbol):
stands for the Greek uppercase letter sigma, which is not to be confused with the sum symbol):![]()
The eigenvectors特征向量 of the covariance matrix represent the principal components (the directions of maximum variance ), whereas the corresponding eigenvalues will define their magnitude. In the case of the Wine dataset, we would obtain 13 eigenvectors and eigenvalues from the 13×13 -dimensional covariance matrix.
Now, let's obtain the eigenpairs of the covariance matrix. As we surely remember from our introductory linear algebra or calculus classes, an eigen vector ![]() satisfies the following condition:
satisfies the following condition:![]()
Here, ![]() is a scalar: the eigenvalue(Lagrange multiplier). Since the manual computation of eigenvectors and eigenvalues is a somewhat tedious and elaborate复杂 task, we will use the linalg.eig function from NumPy to obtain the eigenpairs of the Wine covariance matrix:
is a scalar: the eigenvalue(Lagrange multiplier). Since the manual computation of eigenvectors and eigenvalues is a somewhat tedious and elaborate复杂 task, we will use the linalg.eig function from NumPy to obtain the eigenpairs of the Wine covariance matrix:
import numpy as np
cov_mat = np.cov(X_train_std.T) #the covariance matrix
eigen_vals, eigen_vecs = np.linalg.eig( cov_mat )
print('\nEigenValues \n%s' % eigen_vals![]()
Using the numpy.cov function, we computed the covariance matrix of the standardized training dataset. Using the linalg.eig function, we performed the eigen decomposition, which yielded a vector (eigen_vals) consisting of 13 eigenvalues and the corresponding eigenvectors stored as columns in a 13 x 13-dimensional matrix (eigen_vecs).
####################
Note
The numpy.linalg.eig function was designed to operate on both symmetric and non-symmetric square matrices. However, you may find that it returns complex(复合) eigenvalues in certain cases.
A related function, numpy.linalg.eigh, has been implemented to decompose Hermetian matrices, which is a numerically more stable approach to work with symmetric matrices such as the covariance matrix; numpy.linalg.eigh always returns real eigenvalues.
###################
Total and explained variance
Since we want to reduce the dimensionality of our dataset by compressing it onto a new feature subspace, we only select the subset of the eigenvectors (principal components) that contains most of the information (variance方差最大). The eigenvalues define the magnitude of the eigenvectors(特征值的大小决定了特征向量的重要性), so we have to sort the eigenvalues by decreasing magnitude; we are interested in the top k eigenvectors based on the values of their corresponding eigenvalues. But before we collect those k most informative eigenvectors, let us plot the variance explained ratios of the eigenvalues. The variance explained ratio(方差解释比率或者方差贡献率) of an eigenvalue![]() is simply the fraction of an eigenvalue
is simply the fraction of an eigenvalue![]() and the total sum of the eigenvalues:
and the total sum of the eigenvalues:![]()
Using the NumPy cumsum function, we can then calculate the cumulative sum of explained variances, which we will then plot via Matplotlib's step function:
tot = sum(eigen_vals) #the total sum of the eigenvalues
var_exp = [ (i/tot) for i in sorted(eigen_vals, reverse=True) ] #The variance explained ratio of an eigenvalue
cum_var_exp = np.cumsum( var_exp ) #the cumulative sum of explained variances
import matplotlib.pyplot as plt
#13 eigenvalues
plt.bar( range(1,14), var_exp, alpha=0.5, align="center", label="individual explained variance")
#where="mid": Steps occur half-way between the *x* positions.
plt.step( range(1,14), cum_var_exp, where="mid", label="cumulative explained variance")
plt.ylabel("Explained variance ratio")
plt.xlabel("Principal component index")
plt.legend(loc="best")
![]()
The resulting plot indicates that the first principal component alone accounts for approximately 40 percent of the variance. Also, we can see that the first two principal components combined explain almost 60 percent of the variance in the dataset.
Although the explained variance plot reminds us of the feature importance values that we computed in Chapter 4, Building Good Training Sets – Data Preprocessing, via random forests, we should remind ourselves that PCA is an unsupervised method, which means that information about the class labels is ignored. Whereas a random forest uses the class membership information to compute the node impurities, variance measures the spread of values along a feature axis.
Feature transformation
After we have successfully decomposed the covariance matrix into eigenpairs(eigenvector and eigenvalue), let's now proceed with the last three steps to transform the Wine dataset onto the new principal component axes. The remaining steps we are going to tackle in this section are the following ones:
- Select k eigenvectors, which correspond to the k largest eigenvalues, where k is the dimensionality of the new feature subspace (
 ).
). - Construct a projection matrix W from the "top" k eigenvectors.
- Transform the d-dimensional input dataset X using the projection matrix W to obtain the new k-dimensional feature subspace.
Or, in less technical terms, we will sort the eigenpairs by descending order of the eigenvalues, construct a projection matrix from the selected eigenvectors, and use the projection matrix to transform the data onto the lower-dimensional subspace.
We start by sorting the eigenpairs by decreasing order of the eigenvalues:
# Make a list of (eigenvalue, eigenvector) tuples
eigen_pairs = [ (np.abs(eigen_vals[i]), eigen_vecs[:,i]) for i in range(len(eigen_vals)) ]
eigen_pairs.sort(reverse=True)Next, we collect the two eigenvectors that correspond to the two largest eigenvalues, to capture about 60 percent of the variance in this dataset. Note that we only chose two eigenvectors for the purpose of illustration, since we are going to plot the data via a two-dimensional scatter plot later in this subsection. In practice, the number of principal components has to be determined by a trade-off between computational efficiency and the performance of the classifier:
eigen_pairs[:2]![]()
#eigen_pairs:[(eigen_value, eigen_vector(array type)),
# (eigen_value, eigen_vector),...]
#eigen_pairs[0][1]: get first eigenvector or first principal component
w = np.hstack((eigen_pairs[0][1][:, np.newaxis],#[:, np.newaxis]:the principal component as 1st column
eigen_pairs[1][1][:, np.newaxis] #[:, np.newaxis]:the principal component as 2nd column
)
)#( ([1st column], [second column]) )
print('Matrix W:\n', w)![]()
By executing the preceding code, we have created a 13 x 2-dimensional projection matrix W from the top two eigenvectors.
#########################################
Note
Depending on which version of NumPy and LAPACK(Linear Algebra PACKage) you are using, you may obtain the matrix W with its signs flipped. Please note that this is not an issue; if v is an eigenvector of a (covariance) matrix ![]() , we have:
, we have:
![]()
Here ![]() is our eigenvalue, and
is our eigenvalue, and ![]() - is also an eigenvector(
- is also an eigenvector(![]() and v is a unit vector, note that we are only interested in the direction defined by v) that has the same eigenvalue, since:
and v is a unit vector, note that we are only interested in the direction defined by v) that has the same eigenvalue, since:![]()
#########################################
Using the projection matrix, we can now transform a sample x (represented as a 1 x 13-dimensional row) onto the PCA subspace (the principal components one and two) obtaining ![]() , now a two-dimensional sample vector consisting of two new features:
, now a two-dimensional sample vector consisting of two new features:![]()
X_train_std[0].dot(w)![]()
Similarly, we can transform the entire 124 x 13-dimensional training dataset onto the two principal components by calculating the matrix dot product:![]()
X_train_pca = X_train_std.dot(w)
X_train_std[:3]![]()
X_train_pca[:3]![]()
cp5_Compressing Data via Dimensionality Reduction_feature extraction_PCA_LDA_convergence_kernel PCA : https://blog.csdn.net/Linli522362242/article/details/105196037
###########################################################
In the previous section, we visualized the data distributions of the Housing dataset variables in the form of histograms and scatterplots. Next, we will create a correlation matrix to quantify量化 and summarize linear relationships between variables https://blog.csdn.net/Linli522362242/article/details/103387527. A correlation matrix is closely related to the covariance matrix that we covered in the section Unsupervised dimensionality reduction via principal component analysis in cp5_Compressing Data via Dimensionality Reduction_feature extraction_PCA_LDA_convergence_kernel PCA https://blog.csdn.net/Linli522362242/article/details/105196037. We can interpret the correlation matrix as being a rescaled version of the covariance matrix. In fact, the correlation matrix is identical to a covariance matrix computed from standardized features.
The correlation matrix相关系数矩阵 is a square matrix that contains the Pearson product-moment correlation coefficient 皮尔逊积矩相关系数 (often abbreviated as Pearson's r), which measures the linear dependence between pairs of features. The correlation coefficients are in the range –1 to 1. Two features have a perfect positive correlation if r = 1, no correlation if r = 0, and a perfect negative correlation if r = –1. As mentioned previously, Pearson's correlation coefficient can simply be calculated as the covariance between two features, x and y (numerator), divided by the product of their standard deviations (denominator):
( Pearson product-moment correlation coefficient )Pearson's r : 
# the covariance协方差 between two features ![]() and
and ![]() on the population level can be calculated via the following equation
on the population level can be calculated via the following equation![]()
Here, denotes the mean of the corresponding feature, ![]() is the covariance between the features x and y, and
is the covariance between the features x and y, and ![]() and
and ![]() are the features' standard deviations.
are the features' standard deviations.
Covariance versus correlation for standardized features
We can show that the covariance between a pair of standardized features is, in fact, equal to their linear correlation coefficient. To show this, let's first standardize the features x and y to obtain their z-scores, which we will denote as ′ and ′, respectively: 
Remember that we compute the (population) covariance between two features as follows: ![]()
Since standardization centers a feature variable at mean zero, we can now calculate the covariance between the scaled features as follows: 
Through re-substitution(  ), we then get the following result:
), we then get the following result:
Finally, we can simplify this equation as follows:  ( correlation coefficient formula)
( correlation coefficient formula)
In the following code example, we will use NumPy's corrcoef function on the five feature columns that we previously visualized in the scatterplot matrix, and we will use MLxtend's heatmap function to plot the correlation matrix array as a heat map:
from mlxtend.plotting import heatmap
import numpy as np
cm = np.corrcoef( df[cols].values.T ) # df[cols].values.T : (n_instances, n_features) ==> (n_features, n_instances)
hm = heatmap(cm, row_names=cols, column_names=cols)
plt.show()As you can see in the resulting figure, the correlation matrix provides us with another useful summary graphic that can help us to select features based on their respective linear correlations
![]()
To fit a linear regression model, we are interested in those features that have a high correlation with our target variable, MEDV. Looking at the previous correlation matrix, we can see that our target variable, MEDV, shows the largest correlation with the LSTAT variable (-0.74); however, as you might remember from inspecting the scatterplot matrix, there is a clear nonlinear relationship between LSTAT and MEDV![]() . On the other hand, the correlation between RM and MEDV is also relatively high (0.70). Given the linear relationship between these two variables that we observed in the scatterplot
. On the other hand, the correlation between RM and MEDV is also relatively high (0.70). Given the linear relationship between these two variables that we observed in the scatterplot , RM seems to be a good choice for an exploratory variable to introduce the concepts of a simple linear regression model in the following section.
, RM seems to be a good choice for an exploratory variable to introduce the concepts of a simple linear regression model in the following section.
Implementing an ordinary least squares linear regression model
At the beginning, it was mentioned that linear regression can be understood as obtaining the best-fitting straight line through the examples of our training data. However, we have neither defined the term best-fitting nor have we discussed the different techniques of fitting such a model. In the following subsections, we will fill in the missing pieces of this puzzle using the ordinary least squares (OLS) method (sometimes also called linear least squares) to estimate the parameters of the linear regression line that minimizes the sum of the squared vertical distances (residuals or errors) to the training examples.
Solving regression for regression parameters with gradient descent
Consider our implementation of the Adaptive Linear Neuron (Adaline) from cp02_TrainingSimpleMachineLearningAlgorithmsForClassification_meshgrid_ravel_contourf_OvA_GradientDecent https://blog.csdn.net/Linli522362242/article/details/96429442. You will remember that the artificial neuron uses a linear activation function. Also, we defined a cost function, J(w), which we minimized to learn the weights(![]() ) via optimization algorithms
) via optimization algorithms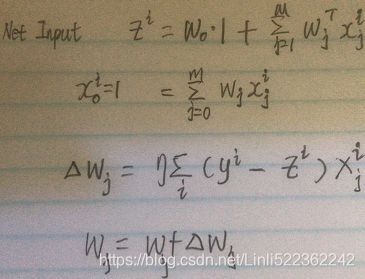 , such as gradient descent (GD) and stochastic gradient descent (SGD, the step size is determined by the value of the learning rate). This cost function in Adaline is the sum of squared errors (SSE), which is identical to the cost function that we use for OLS:
, such as gradient descent (GD) and stochastic gradient descent (SGD, the step size is determined by the value of the learning rate). This cost function in Adaline is the sum of squared errors (SSE), which is identical to the cost function that we use for OLS:
Here, ̂ is the predicted value ̂ = ![]() (note that the term
(note that the term![]() is just used for convenience to derive the update rule of GD). Essentially, OLS regression can be understood as Adaline without the unit step function so that we obtain continuous target values instead of the class labels -1 and 1. To demonstrate this, let's take the GD implementation of Adaline from cp02_TrainingSimpleMachineLearningAlgorithmsForClassification_meshgrid_ravel_contourf_OvA_GradientDecent https://blog.csdn.net/Linli522362242/article/details/96429442, and remove the unit step function to implement our first linear regression model:
is just used for convenience to derive the update rule of GD). Essentially, OLS regression can be understood as Adaline without the unit step function so that we obtain continuous target values instead of the class labels -1 and 1. To demonstrate this, let's take the GD implementation of Adaline from cp02_TrainingSimpleMachineLearningAlgorithmsForClassification_meshgrid_ravel_contourf_OvA_GradientDecent https://blog.csdn.net/Linli522362242/article/details/96429442, and remove the unit step function to implement our first linear regression model:
class LinearRegressionGD(object):
def __init__(self, eta=0.001, n_iter=20):
self.eta = eta
self.n_iter = n_iter
def net_input(self, X):
return np.dot(X, self.w_[1:]) + self.w_[0] # result's shape: (num_instances in X) ==(X.shape[0])
def fit(self, X,y):
self.w_ = np.zeros(1+X.shape[1]) # X.shape[1] : num_features
# OR
# rgen = np.random.RandomState(self.random_state) #1+ num_features
# self.w_ = rgen.normal( loc=0.0, scale=0.01, size=1+X.shape[1] )
self.cost_ = []
for i in range( self.n_iter ):
output = self.net_input(X) # single column matrix
errors = (y-output)
self.w_[1:] += self.eta * X.T.dot(errors) # X.T : (num_features, num_instances)
self.w_[0] += self.eta * errors.sum()
cost = (errors**2).sum() /2.0
self.cost_.append(cost)
return self
def predict(self,X):
return self.net_input(X)Weight updates with gradient descent
If you need a refresher about how the weights are updated—taking a step in the opposite direction of the gradient—please revisit the Adaptive linear neurons and the convergence of learning section in Chapter 2, Training Simple Machine Learning Algorithms for Classification.
To see our LinearRegressionGD regressor in action, let's use the RM (number of rooms) variable from the Housing dataset as the explanatory variable and train a model that can predict MEDV (house prices). Furthermore, we will standardize the variables for better convergence of the GD algorithm. The code is as follows:
X = df[['RM']].values # 2D array : 506 rows × 1 columns
y = df['MEDV'].values # 1D array
from sklearn.preprocessing import StandardScaler
sc_x = StandardScaler()
sc_y = StandardScaler()
X_std = sc_x.fit_transform(X)
y_std = sc_y.fit_transform(y[:, np.newaxis]).flatten() # y[:, np.newaxis] since fit_transform will take 2D array
# flatten() to 1D again
lr = LinearRegressionGD()
lr.fit(X_std, y_std)![]()
Notice the workaround regarding y_std, using np.newaxis and flatten. Most transformers in scikit-learn expect data to be stored in two-dimensional arrays. In the previous code example, the use of np.newaxis in y[:, np.newaxis] added a new dimension to the array. Then, after StandardScaler returned the scaled variable, we converted it back to the original one-dimensional array representation using the flatten() method for our convenience.
We discussed in Chapter 2, Training Simple Machine Learning Algorithms for Classification https://blog.csdn.net/Linli522362242/article/details/96429442, that it is always a good idea to plot the cost as a function of the number of epochs (complete iterations) over the training dataset when we are using optimization algorithms, such as GD, to check that the algorithm converged to a cost minimum (here, a global cost minimum):
plt.plot( range(1, lr.n_iter+1),
lr.cost_ )
plt.ylabel('SSE')
plt.xlabel('Epoch')
plt.grid(False)
plt.show()As you can see in the following plot, the GD algorithm converged after the fifth epoch:
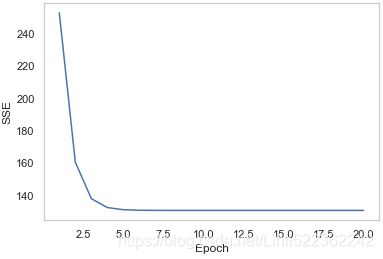
Next, let's visualize how well the linear regression line fits the training data. To do so, we will define a simple helper function that will plot a scatterplot of the training examples and add the regression line:
def lin_regplot(X,y, model):
plt.scatter( X,y, c='steelblue', edgecolor='white', s=70 )
plt.plot(X, model.predict(X), color='black', lw=2 )
return # or # return NoneNow, we will use this lin_regplot function to plot the number of rooms against the house price:
lin_regplot(X_std, y_std, lr)
plt.xlabel('Average number of rooms [RM] (standardized)' )
plt.ylabel('Price in $1000s [MEDV] (standardized)')
plt.show()As you can see in the following plot, the linear regression line reflects the general trend that house prices tend to increase with the number of rooms:
Although this observation makes sense, the data also tells us that the number of rooms does not explain house prices very well in many cases. Later, we will discuss how to quantify量化 the performance of a regression model. Interestingly, we can also observe that several data points lined up at y = 3, which suggests that the prices may have been clipped(这意味着房价被限定了上界). In certain applications, it may also be important to report the predicted outcome variables on their original scale. To scale the predicted price outcome back onto the Price in $1000s axis, we can simply apply the inverse_transform method of the StandardScaler:
num_rooms_std = sc_x.transform( np.array([[5.0]]) )
price_std = lr.predict( num_rooms_std )
print('Price in $1000s: %.3f' % sc_y.inverse_transform(price_std) )![]()
In this code example, we used the previously trained linear regression model to predict the price of a house with five rooms. According to our model, such a house will be worth $10,840.
As a side note, it is also worth mentioning that we technically don't have to update the weights of the intercept if we are working with standardized variables, since the y axis intercept is always 0 in those cases. We can quickly confirm this by printing the weights:
print('Slope: %.3f' % lr.w_[1])
print('Intercept: %.3f' % lr.w_[0])Estimating the coefficient of a regression model via scikit-learn
In the previous section, we implemented a working model for regression analysis; however, in a real-world application, we may be interested in more efficient implementations. For example, many of scikit-learn's estimators for regression make use of the least squares implementation in SciPy (scipy.linalg.lstsq), which in turn uses highly optimized code optimizations based on the Linear Algebra Package (LAPACK). The linear regression implementation in scikit-learn also works (better) with unstandardized variables, since it does not use (S)GD-based optimization, so we can skip the standardization step:
from sklearn.linear_model import LinearRegression
slr = LinearRegression()
slr.fit(X,y)
y_pred = slr.predict(X)
print('Slope: %.3f' % slr.coef_[0])
print('Intercept: %.3f' % slr.intercept_)As you can see from executing this code, scikit-learn's LinearRegression model, fitted with the unstandardized RM and MEDV variables, yielded different model coefficients, since the features have not been standardized. However, when we compare it to our GD implementation by plotting MEDV against RM, we can qualitatively see that it fits the data similarly well:
lin_regplot(X,y,slr)
plt.xlabel('Average number of rooms [RM]')
plt.ylabel('Price in %1000s [MEDV]')
plt.grid(False)
plt.show()For instance, we can see that the overall result looks identical to our GD implementation:
Analytical solutions of linear regression
As an alternative to using machine learning libraries, there is also a closed-form solution for solving OLS involving a system of linear equations that can be found in most introductory statistics textbooks:![]()
OR https://blog.csdn.net/Linli522362242/article/details/104005906
![]() 04_TrainingModels_Normal Equation(正态方程,正规方程) Derivation_Gradient Descent_Polynomial Regressionhttps://blog.csdn.net/Linli522362242/article/details/104005906
04_TrainingModels_Normal Equation(正态方程,正规方程) Derivation_Gradient Descent_Polynomial Regressionhttps://blog.csdn.net/Linli522362242/article/details/104005906
Equation 4-3. MSE cost function for a Linear Regression model ![]()
solve Normal Equation( has a closed-form solution) ==> find the value of θ (Minimum ![]() ==0
==0 ![]() )==> minimizes MSE cost function ==> minimizes the RMSE
)==> minimizes MSE cost function ==> minimizes the RMSE
We can implement it in Python as follows:
# adding a column vector of "ones"
Xb = np.hstack(( np.ones( (X.shape[0], 1) ), X ))
w = np.zeros(X.shape[1])
z = np.linalg.inv( np.dot(Xb.T, Xb) )
w = np.dot(z, np.dot(Xb.T, y))
print('Slope: %.3f' % w[1])
print('Intercept: %.3f' % w[0])![]()
The advantage of this method is that it is guaranteed to find the optimal solution analytically. However, if we are working with very large datasets, it can be computationally too expensive to invert the matrix in this formula (sometimes also called the normal equation), or the matrix containing the training examples may be singular (non-invertible), which is why we may prefer iterative methods in certain cases.
If you are interested in more information on how to obtain normal equations, take a look at Dr. Stephen Pollock's chapter The Classical Linear Regression Model from his lectures
at the University of Leicester, which is available for free at:http://www.le.ac.uk/users/dsgp1/COURSES/MESOMET/ECMETXT/06mesmet.pdf.
Also, if you want to compare linear regression solutions obtained via GD, SGD, the closed-form solution, QR factorization, and singular vector decomposition, you can use the LinearRegression class implemented in MLxtend (http://rasbt.github.io/mlxtend/user_guide/regressor/LinearRegression/), which lets users toggle between these options. Another great library to recommend for regression modeling in Python is Statsmodels, which implements more advanced linear regression models, as illustrated at https://www.statsmodels.org/stable/examples/index.html#regression.
Fitting a robust regression model using RANSAC
Linear regression models can be heavily impacted by the presence of outliers. In certain situations, a very small subset of our data can have a big effect on the estimated model coefficients. There are many statistical tests that can be used to detect outliers, which are beyond the scope of the book. However, removing outliers always requires our own judgment as data scientists as well as our domain knowledge.
As an alternative to throwing out outliers, we will look at a robust method of regression using the RANdom SAmple Consensus[kənˈsensəs]一致 (RANSAC) algorithm, which fits a regression model to a subset of the data, the so-called inliers.
We can summarize the iterative RANSAC algorithm as follows:
- 1. Select a random number of examples to be inliers and fit the model.
- 2. Test all other data points against the fitted model and add those points that fall within a user-given tolerance to the inliers.
- 3. Refit the model using all inliers.
- 4. Estimate the error of the fitted model versus the inliers.
- 5. Terminate the algorithm if the performance meets a certain user-defined threshold or if a fixed number of iterations were reached; go back to step 1 otherwise.
Let's now use a linear model in combination with the RANSAC algorithm as implemented in scikit-learn's RANSACRegressor class:
###############################################################
sklearn.linear_model.RANSACRegressor
class sklearn.linear_model.RANSACRegressor(base_estimator=None, *, min_samples=None, residual_threshold=None, is_data_valid=None, is_model_valid=None, max_trials=100,
max_skips=inf, stop_n_inliers=inf, stop_score=inf, stop_probability=0.99, loss='absolute_loss', random_state=NoneRANSAC (RANdom SAmple Consensus) algorithm.
RANSAC is an iterative algorithm for the robust estimation of parameters from a subset of inliers from the complete data set.
min_samples : int (>= 1) or float ([0, 1]), optional
Minimum number of samples chosen randomly from original data. Treated as an absolute number of samples for min_samples >= 1, treated as a relative number ceil(min_samples * X.shape[0]) for min_samples < 1. This is typically chosen as the minimal number of samples necessary to estimate the given base_estimator. By default a sklearn.linear_model.LinearRegression() estimator is assumed and min_samples is chosen as X.shape[1] + 1.
residual_threshold : float, optional
Maximum residual for a data sample to be classified as an inlier. By default the threshold is chosen as the MAD (median absolute deviation) of the target values y.
max_trials : int, optional
Maximum number of iterations for random sample selection.
loss : string, callable, optional, default “absolute_loss”
String inputs, “absolute_loss” and “squared_loss” are supported which find the absolute loss and squared loss per sample respectively.
If loss is a callable, then it should be a function that takes two arrays as inputs, the true and predicted value and returns a 1-D array with the i-th value of the array corresponding to the loss on X[i].
If the loss on a sample is greater than the residual_threshold, then this sample is classified as an outlier.
Attributes:
inlier_mask_ : bool array of shape [n_samples]
Boolean mask of inliers classified as True.
###############################################################
04_TrainingModels_Normal Equation(正态方程,正规方程) Derivation_Gradient Descent_Polynomial Regression : https://blog.csdn.net/Linli522362242/article/details/104005906
![]()
![]()
from sklearn.linear_model import RANSACRegressor
ransac = RANSACRegressor( LinearRegression(),
max_trials = 100,
min_samples = 50,
loss = 'absolute_loss',
residual_threshold=5.0,
random_state=0 )
ransac.fit(X,y)We set the maximum number of iterations of the RANSACRegressor to 100, and using min_samples=50, we set the minimum number of the randomly chosen training examples to be at least 50. Using 'absolute_loss' as an argument for the loss parameter, the algorithm computes absolute vertical distances between the fitted line and the training examples. By setting the residual_threshold parameter to 5.0, we only allow training examples to be included in the inlier set if their vertical distance to the fitted line(model) is within 5 distance units, which works well on this particular dataset. By default, scikit-learn uses the MAD estimate to select the inlier threshold, where MAD stands for the median absolute deviation of the target values, y. However, the choice of an appropriate value for the inlier threshold is problem-specific, which is one disadvantage of RANSAC. Many different approaches have been developed in recent years to select a good inlier threshold automatically. You can find a detailed discussion in Automatic Estimation of the Inlier Threshold in Robust Multiple Structures Fitting, R. Toldo, A. Fusiello's, Springer, 2009 (in Image Analysis and Processing–ICIAP 2009, pages: 123-131).
Once we have fitted the RANSAC model, let's obtain the inliers and outliers from the fitted RANSAC-linear regression model and plot them together with the linear fit:
inlier_mask = ransac.inlier_mask_
outlier_mask = np.logical_not( inlier_mask )
line_X = np.arange(3,10,1) # since the previous plot############################
line_y_ransac = ransac.predict( line_X[:, np.newaxis] )
plt.scatter( X[inlier_mask], y[inlier_mask],
c='steelblue', edgecolor = 'white',
marker='o', label='Inliers')
plt.scatter( X[outlier_mask], y[outlier_mask],
c='limegreen', edgecolor='white',
marker='s', label='Outliers')
plt.plot(line_X, line_y_ransac, color='black', lw=2)
plt.xlabel('Average number of rooms[RM]')
plt.ylabel('Price in $1000s [MEDV]')
plt.legend(loc='upper left')
plt.show()As you can see in the following scatterplot, the linear regression model was fitted on the detected set of inliers, which are shown as circles:
When we print the slope and intercept of the model by executing the following code, the linear regression line will be slightly different from the fit that we obtained in the previous section without using RANSAC:
print('Slope: %.3f' % ransac.estimator_.coef_[0])
print('Intercept: %.3f' % ransac.estimator_.intercept_)Using RANSAC, we reduced the potential effect of the outliers in this dataset, but we don't know whether this approach will have a positive effect on the predictive performance for unseen data or not. Thus, in the next section, we will look at different approaches for evaluating a regression model, which is a crucial part of building systems for predictive modeling.
Evaluating the performance of linear regression models
In the previous section, you learned how to fit a regression model on training data. However, you discovered in previous chapters that it is crucial to test the model on data that it hasn't seen during training to obtain a more unbiased estimate of its generalization performance.
As you will remember from cp6_Model Eval_Confusion_Hyperpara Tuning_pipeline_variance_bias_ validation_learning curve_strength https://blog.csdn.net/Linli522362242/article/details/109560084, we want to split our dataset into separate training and test datasets, where we will use the former to fit the model and the latter to evaluate its performance on unseen data to estimate the generalization performance. Instead of proceeding with the simple regression model, we will now use all variables in the dataset and train a multiple regression model:
from sklearn.model_selection import train_test_split
X = df.iloc[:, :-1].values #exclude the last feature(or the last column)
y = df['MEDV'].values
X_train, X_test, y_train, y_test = train_test_split(
X, y, test_size=0.3, random_state=0
)
slr = LinearRegression()
slr.fit(X_train, y_train)
y_train_pred = slr.predict(X_train)
y_test_pred = slr.predict(X_test)Since our model uses multiple explanatory variables, we can't visualize the linear regression line (or hyperplane, to be precise) in a two-dimensional plot, but we can plot the residuals (the differences or vertical distances between the actual and predicted values) versus the predicted values to diagnose our regression model. Residual plots are a commonly used graphical tool for diagnosing regression models. They can help to detect nonlinearity and outliers, and check whether the errors are randomly distributed.
Using the following code, we will now plot a residual plot where we simply subtract the true target variables from our predicted responses:
plt.figure(figsize=(10,8))
# residuals
plt.scatter( y_train_pred, y_train_pred - y_train,
c='steelblue', marker='o', edgecolors='white',
label = 'Training data' )
plt.scatter( y_test_pred, y_test_pred - y_test,
c='limegreen', marker='s', edgecolors='white',
label = 'Test data' )
plt.xlabel('Predicted values')
plt.ylabel('Residuals')
plt.legend( loc='upper left')
plt.hlines(y=0, xmin=-10, xmax=50, color='black', lw=2)
plt.xlim([-10,50])
plt.tight_layout()
plt.show()After executing the code, we should see a residual plot with a line passing through the x axis origin, as shown here: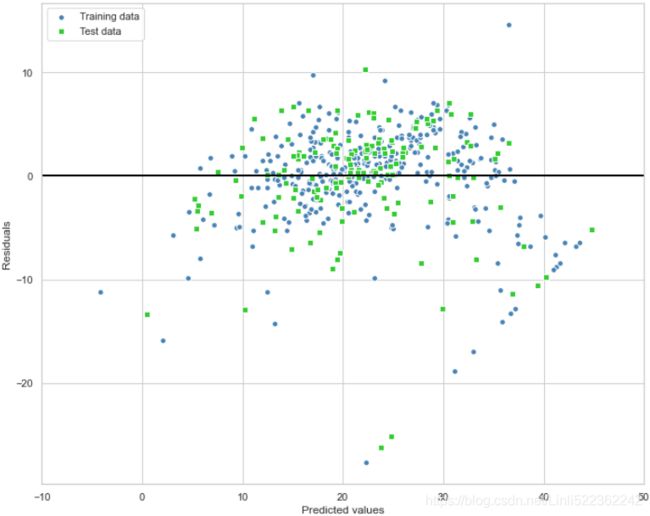
In the case of a perfect prediction, the residuals would be exactly zero, which we will probably never encounter in realistic and practical applications. However, for a good regression model, we would expect the errors to be randomly distributed and the residuals to be randomly scattered around the centerline. If we see patterns in a residual plot, it means that our model is unable to capture some explanatory information, which has leaked into the residuals, as you can slightly see in our previous residual plot. Furthermore, we can also use residual plots to detect outliers, which are represented by the points with a large deviation from the centerline.
Another useful quantitative measure of a model's performance is the so-called mean squared error (MSE), which is simply the averaged value of the SSE cost that we minimized to fit the linear regression model. The MSE is useful for comparing different regression models or for tuning their parameters via grid search and crossvalidation, as it normalizes the SSE by the sample size: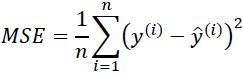
Let's compute the MSE of our training and test predictions:
from sklearn.metrics import mean_squared_error
print('MSE train: %.3f, test: %.3f' % ( mean_squared_error(y_train, y_train_pred),
mean_squared_error(y_test, y_test_pred)
) )![]()
You can see that the MSE on the training dataset is 19.96, and the MSE on the test dataset is much larger, with a value of 27.20, which is an indicator that our model is overfitting the training data in this case. However, please be aware that the MSE is unbounded in contrast to the classification accuracy, for example. In other words, the interpretation of the MSE depends on the dataset and feature scaling. For example, if the house prices were presented as multiples of 1,000 (with the K suffix), the same model would yield a lower MSE compared to a model that worked with unscaled features. To further illustrate this point, ($10K − 15K)^2 < ($10,000 − $15,000)^2 .
Thus, it may sometimes be more useful to report the coefficient of determination决定系数 (![]() ) , which can be understood as a standardized version of the MSE, for better interpretability of the model's performance. Or, in other words,
) , which can be understood as a standardized version of the MSE, for better interpretability of the model's performance. Or, in other words, ![]() is the fraction of response variance响应方差的分数 that is captured by the model. The
is the fraction of response variance响应方差的分数 that is captured by the model. The ![]() value is defined as:
value is defined as: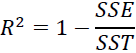 OR
OR 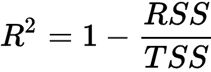
Here, SSE is the sum of squared errors(OR the sum of squared of residuals)
and SST is the total sum of squares: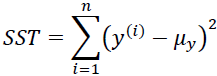
In other words, SST is simply the variance of the response.
Let's quickly show that ![]() is indeed just a rescaled version of the MSE:
is indeed just a rescaled version of the MSE: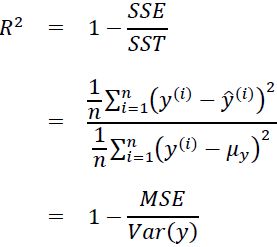 #
# 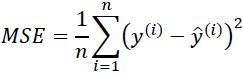 # rescaled y :
# rescaled y : ![]()
For the training dataset, the ![]() is bounded between 0 and 1, but it can become negative for the test dataset. If
is bounded between 0 and 1, but it can become negative for the test dataset. If ![]() = 1 , the model fits the data perfectly with a corresponding MSE = 0(since Var(y)>0).
= 1 , the model fits the data perfectly with a corresponding MSE = 0(since Var(y)>0).
Evaluated on the training data, the ![]() of our model is 0.765, which doesn't sound too bad. However, the
of our model is 0.765, which doesn't sound too bad. However, the ![]() on the test dataset is only 0.673, which we can compute by executing the following code:
on the test dataset is only 0.673, which we can compute by executing the following code:
from sklearn.metrics import r2_score
print('R^2 train: %.3f, test: %.3f' % ( r2_score(y_train, y_train_pred),
r2_score(y_test, y_test_pred)
) )![]()
Coefficient of determination, in statistics, ![]() (or r^2), a measure that assesses the ability of a model to predict or explain an outcome in the linear regression setting. More specifically, R2
(or r^2), a measure that assesses the ability of a model to predict or explain an outcome in the linear regression setting. More specifically, R2![]() indicates the proportion of the variance方差 in the dependent variable 因变量(Y) that is predicted or explained by linear regression and the predictor variable (X, also known as the independent variable自变量).
indicates the proportion of the variance方差 in the dependent variable 因变量(Y) that is predicted or explained by linear regression and the predictor variable (X, also known as the independent variable自变量).
The coefficient of determination shows only association. As with linear regression, it is impossible to use ![]() to determine whether one variable causes the other. In addition, the coefficient of determination shows only the magnitude of the association, not whether that association is statistically significant.
to determine whether one variable causes the other. In addition, the coefficient of determination shows only the magnitude of the association, not whether that association is statistically significant.
In general, a high R2 value indicates that the model is a good fit for the data, although interpretations of fit depend on the context of analysis. An R2 of 0.35, for example, indicates that 35 percent of the variation变动,差异 in the outcome has been explained just by predicting the outcome using the covariates(协变量,![]() ) included in the model表明仅通过使用模型中包含的协变量, 预测结果即可解释结果差异的35%. That percentage
) included in the model表明仅通过使用模型中包含的协变量, 预测结果即可解释结果差异的35%. That percentage![]() might be a very high portion of variation to predict in a field such as the social sciences; in other fields, such as the physical sciences, one would expect R2 to be much closer to 100 percent. The theoretical minimum R2 is 0. However, since linear regression is based on the best possible fit, R2 will always be greater than zero, even when the predictor(X) and outcome variables(Y) bear no relationship to one another.
might be a very high portion of variation to predict in a field such as the social sciences; in other fields, such as the physical sciences, one would expect R2 to be much closer to 100 percent. The theoretical minimum R2 is 0. However, since linear regression is based on the best possible fit, R2 will always be greater than zero, even when the predictor(X) and outcome variables(Y) bear no relationship to one another.![]() OR
OR 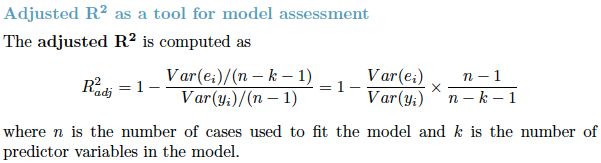
R2 increases when a new predictor variable is added to the model, even if the new predictor is not associated with the outcome![]() . To account for that effect, the adjusted R2 (typically denoted with a bar over the R in R2) incorporates the same information as the usual
. To account for that effect, the adjusted R2 (typically denoted with a bar over the R in R2) incorporates the same information as the usual ![]() but then also penalizes for the number(k) of predictor variables included in the model. As a result, R2 increases as new predictors are added to a multiple linear regression model, but the adjusted R2 increases only if the increase in R2 is greater than one would expect from chance alone
but then also penalizes for the number(k) of predictor variables included in the model. As a result, R2 increases as new predictors are added to a multiple linear regression model, but the adjusted R2 increases only if the increase in R2 is greater than one would expect from chance alone![]() 仅当新项对模型的改进超出偶然的预期时,the adjusted R2 才会增加. It decreases when a predictor improves the model by less than expected by chance.
仅当新项对模型的改进超出偶然的预期时,the adjusted R2 才会增加. It decreases when a predictor improves the model by less than expected by chance.![]() In such a model, the adjusted R2 is the most realistic estimate of the proportion of the variation that is predicted by the covariates included in the model
In such a model, the adjusted R2 is the most realistic estimate of the proportion of the variation that is predicted by the covariates included in the model
Using regularized methods for regression
As we discussed in cp3 A Tour of ML Classifiers_stratify_bincount_likelihood_logistic regression_odds ratio_decay_L2 https://blog.csdn.net/Linli522362242/article/details/96480059, regularization(e.g. L2 shrinkage or weight decay) is one approach to tackling the problem of overfitting by adding additional information, and thereby shrinking the parameter values of the model to induce a penalty against complexity. The most popular approaches to regularized linear regression are the so-called Ridge Regression, least absolute shrinkage and selection operator (LASSO), and elastic Net.
Ridge Regression is an L2 penalized model where we simply add the squared sum of the weights to our least-squares cost function :https://blog.csdn.net/Linli522362242/article/details/104070847 and https://blog.csdn.net/Linli522362242/article/details/108230328
minus Predicted_y : 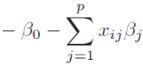
 OR
OR ![]() OR
OR ![]()
Here:
#######################################################################################
Note(![]() is different with MSE(Mean Square of Error)
is different with MSE(Mean Square of Error) ![]() since
since ![]() ): is different with MSE(Mean Square of Error)
): is different with MSE(Mean Square of Error) ![]() since
since ![]() ):
):
#######################################################################################
By increasing the value of hyperparameter , we increase the regularization strength and thereby shrink the weights of our model. Please note that we don't regularize the intercept term, ![]() .
.
##################################Here ![]() ==W
==W
将 (3.41) 的准则写成矩阵形式 ![]()
可以简单地看出岭回归的解为 ![]() (本质在自变量信息矩阵的主对角线元素上人为地加入一个非负因子)
(本质在自变量信息矩阵的主对角线元素上人为地加入一个非负因子)
##################################
An alternative approach that can lead to sparse models is LASSO. Depending on the regularization strength, certain weights can become zero, which also makes LASSO useful as a supervised feature selection technique: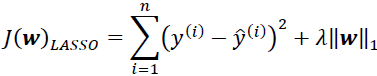 OR
OR ![]()
Here, the L1 penalty for LASSO is defined as the sum of the absolute magnitudes of the model weights, as follows:
However, a limitation of LASSO is that it selects at most n features if m > n, where n is the number of training examples. This may be undesirable in certain applications of feature selection. In practice, however, this property of LASSO is often an advantage because it avoids saturated models. Saturation of a model occurs if the number of training examples is equal to the number of features, which is a form of overparameterization过度参数化. As a consequence, a saturated model can always fit the training data perfectly but is merely a form of interpolation插补 and thus is not expected to generalize well.
A compromise between Ridge Regression and LASSO is elastic net, which has an L1 penalty to generate sparsity(Lasso Regression automatically performs feature selection and outputs a sparse model (i.e., with few nonzero feature weights).) and an L2 penalty such that it can be used for selecting more than n features if m > n:
######################################################https://blog.csdn.net/Linli522362242/article/details/104070847![]()
If α = 0 then Ridge Regression is just Linear Regression.
If α(inverse of ) is very large, then all weights end up![]() very close to zero and the result is a flat line going through the data’s mean
very close to zero and the result is a flat line going through the data’s mean
Note how increasing α leads to flatter (i.e., less extreme, more reasonable) predictions; this reduces the model’s variance but increases its bias. https://blog.csdn.net/Linli522362242/article/details/108230328
https://blog.csdn.net/Linli522362242/article/details/108230328
in the preceding figure, we can see that the contour of the cost function touches the L1 diamond at ![]() . Since the contours of an L1 regularized system are sharp, it is more likely that the optimum—that is, the intersection between the ellipses of the cost function and the boundary of the L1 diamond—is located on the axes, which encourages sparsity. The mathematical details of why L1 regularization can lead to sparse solutions are beyond the scope of this book. If you are interested, an excellent section on L2 versus L1 regularization can be found in section 3.4 of The Elements of Statistical Learning, Trevor Hastie, Robert Tibshirani, and Jerome Friedman, Springer.
. Since the contours of an L1 regularized system are sharp, it is more likely that the optimum—that is, the intersection between the ellipses of the cost function and the boundary of the L1 diamond—is located on the axes, which encourages sparsity. The mathematical details of why L1 regularization can lead to sparse solutions are beyond the scope of this book. If you are interested, an excellent section on L2 versus L1 regularization can be found in section 3.4 of The Elements of Statistical Learning, Trevor Hastie, Robert Tibshirani, and Jerome Friedman, Springer.
by increasing the regularization strength via the regularization parameter ![]() , we shrink the weights towards zero and decrease the dependence of our model on the training data. Let's illustrate this concept in the following figure for the L2 penalty term.
, we shrink the weights towards zero and decrease the dependence of our model on the training data. Let's illustrate this concept in the following figure for the L2 penalty term.
![]()
The quadratic L2 regularization term is represented by the shaded ball. Here, our weight coefficients cannot exceed our regularization budget—the combination of the weight coefficients###W=w1, w2, w3...wn### cannot fall outside the shaded area. On the other hand, we still want to minimize the cost function(such as  The term
The term ![]() is just added for our convenience https://blog.csdn.net/Linli522362242/article/details/96480059). Under the penalty constraint, our best effort is to choose the point where the L2 ball intersects with the contours of the unpenalized cost function. The larger the value of the regularization parameter
is just added for our convenience https://blog.csdn.net/Linli522362242/article/details/96480059). Under the penalty constraint, our best effort is to choose the point where the L2 ball intersects with the contours of the unpenalized cost function. The larger the value of the regularization parameter ![]() gets, the faster the penalized cost function
gets, the faster the penalized cost function![]() grows, which leads to a narrower L2 ball
grows, which leads to a narrower L2 ball![]() . For example, if we increase the regularization parameter
. For example, if we increase the regularization parameter![]() towards infinity, the weight coefficients will become effectively zero, denoted by the center of the L2 ball. To summarize the main message of the example: our goal is to minimize the sum of the unpenalized cost function plus the penalty term, which can be understood as adding bias and preferring a simpler model to reduce the variance(try to underfit) in the absence of sufficient training data to fit the model.
towards infinity, the weight coefficients will become effectively zero, denoted by the center of the L2 ball. To summarize the main message of the example: our goal is to minimize the sum of the unpenalized cost function plus the penalty term, which can be understood as adding bias and preferring a simpler model to reduce the variance(try to underfit) in the absence of sufficient training data to fit the model.
######################################################
Those regularized regression models are all available via scikit-learn, and their usage is similar to the regular regression model except that we have to specify the regularization strength via the parameter , for example, optimized via k-fold crossvalidation.
A Ridge Regression model can be initialized via:
from sklearn.linear_model import Ridge
ridge = Ridge(alpha=1.0)#################################################
As with Linear Regression, we can perform Ridge Regression either by computing a closed-form equation or by performing Gradient Descent. The pros and cons are the same. Equation 4-9 shows the closed-form solution (where A is the n × n identity单位 matrix A( A square matrix full of 0s except for 1s on the main diagonal (top-left to bottom-right). ) except with a 0 in the top-left cell, corresponding to the bias term ).
Equation 4-9. Ridge Regression closed-form solution![]()
![]() Note: Normal Equation(a closed-form equation)
Note: Normal Equation(a closed-form equation) ![]()
Here is how to perform Ridge Regression with Scikit-Learn using a closed-form solution (a variant of Equation 4-9 using a matrix factorization technique by André-Louis Cholesky):
from sklearn.linear_model import Ridge
ridge_reg = Ridge(alpha=1, solver="cholesky", random_state=42)
ridge_reg.fit(X,y)
ridge_reg.predict([[1.5]]) ![]()
ridge_reg = Ridge(alpha=1, solver="sag", random_state=42)
ridge_reg.fit(X,y)
ridge_reg.predict([[1.5]])![]()
solver{‘auto’, ‘svd’, ‘cholesky’, ‘lsqr’, ‘sparse_cg’, ‘sag’, ‘saga’}, default=’auto’
Solver to use in the computational routines:
-
‘auto’ chooses the solver automatically based on the type of data.
-
‘svd’ uses a Singular Value Decomposition(https://blog.csdn.net/Linli522362242/article/details/105139547) of X to compute the Ridge coefficients
 . More stable for singular matrices than ‘cholesky’.
. More stable for singular matrices than ‘cholesky’. -
‘cholesky’ uses the standard scipy.linalg.solve function to obtain a closed-form solution.
-
‘sparse_cg’ uses the conjugate gradient solver共轭梯度求解器 as found in scipy.sparse.linalg.cg. As an iterative algorithm, this solver is more appropriate than ‘cholesky’ for large-scale data (possibility to set
tolandmax_iter). -
‘lsqr’ uses the dedicated regularized least-squares routine scipy.sparse.linalg.lsqr. It is the fastest and uses an iterative procedure.
-
‘sag’ uses a Stochastic Average Gradient descent, and ‘saga’ uses its improved, unbiased version named SAGA. Both methods also use an iterative procedure, and are often faster than other solvers when both n_samples and n_features are large. Note that ‘sag’ and ‘saga’ fast convergence is only guaranteed on features with approximately the same scale. You can preprocess the data with a scaler from sklearn.preprocessing.
#################################################
Note that the regularization strength is regulated by the parameter alpha, which is similar to the parameter . Likewise, we can initialize a LASSO regressor from the linear_model submodule:
from sklearn.linear_model import Lasso
lasso = Lasso(alpha=1.0)Lastly, the ElasticNet implementation allows us to vary the L1 to L2 ratio: OR
OR ![]() https://blog.csdn.net/Linli522362242/article/details/104070847
https://blog.csdn.net/Linli522362242/article/details/104070847
from sklearn.linear_model import ElasticNet
elasticNet = ElasticNet( alpha=1.0, l1_ratio=0.5) For example, if we set the l1_ratio to 1.0, the ElasticNet regressor would be equal to LASSO regression. For more detailed information about the
different implementations of linear regression, please see the documentation at http://scikit-learn.org/stable/modules/linear_model.html.
Turning a linear regression model into a curve – polynomial regression
In the previous sections, we assumed a linear relationship between explanatory and response variables. One way to account for the violation of linearity assumption is to use a polynomial regression model by adding polynomial terms:![]()
Here, d denotes the degree of the polynomial. Although we can use polynomial regression to model a nonlinear relationship, it is still considered a multiple linear regression model because of the linear regression coefficients, w. In the following subsections, we will see how we can add such polynomial terms to an existing dataset conveniently and fit a polynomial regression model.
Adding polynomial terms using scikit-learn
We will now learn how to use the PolynomialFeatures transformer class from scikit-learn to add a quadratic term (d = 2) to a simple regression problem with one
explanatory variable. Then, we will compare the polynomial to the linear fit by following these steps:
1. Add a second-degree polynomial term:
X = np.array([258.0, 270.0, 294.0,
320.0, 342.0, 368.0,
396.0, 446.0, 480.0, 586.0])\
[:, np.newaxis]
y = np.array([236.4, 234.4, 252.8,
298.6, 314.2, 342.2,
360.8, 368.0, 391.2,
390.8])from sklearn.preprocessing import PolynomialFeatures
X_new = np.arange(250, 600, 10)[:, np.newaxis] # array([ [250],[260],..., [590] ])
# 2. Fit a simple linear regression model for comparison:
lr = LinearRegression()
lr.fit(X,y)
y_lin_fit = lr.predict(X_new)
# 4. fit quadratic features
pr = LinearRegression()
# So let’s use Scikit-Learn’s PolynomialFeatures class to transform our training data,
# adding the square (2nd-degree polynomial) of each feature in the training set as new features
# (in this case there is just one feature):
quadratic = PolynomialFeatures(degree=2)
X_quad = quadratic.fit_transform(X)
# bias=1 258.0 66,564
# X_quad
# array([[1.00000e+00, 2.58000e+02, 6.65640e+04],
# [1.00000e+00, 2.70000e+02, 7.29000e+04],
# [1.00000e+00, 2.94000e+02, 8.64360e+04],
# [1.00000e+00, 3.20000e+02, 1.02400e+05],
# [1.00000e+00, 3.42000e+02, 1.16964e+05],
# [1.00000e+00, 3.68000e+02, 1.35424e+05],
# [1.00000e+00, 3.96000e+02, 1.56816e+05],
# [1.00000e+00, 4.46000e+02, 1.98916e+05],
# [1.00000e+00, 4.80000e+02, 2.30400e+05],
# [1.00000e+00, 5.86000e+02, 3.43396e+05]])
pr.fit(X_quad, y)
y_quad_fit = pr.predict( quadratic.fit_transform(X_new) )
# 4. Plot the results:
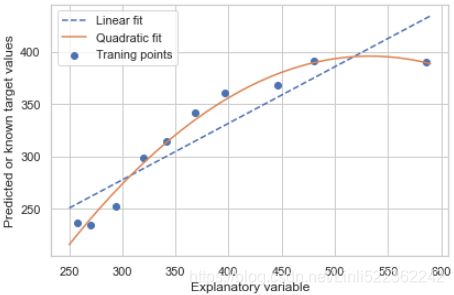
plt.scatter( X,y,label='Traning points' )
plt.plot( X_new, y_lin_fit, label='Linear fit', linestyle='--' )
plt.plot( X_new, y_quad_fit, label='Quadratic fit')
plt.xlabel('Explanatory variable')
plt.ylabel('Predicted or known target values')
plt.legend( loc='upper left')
plt.tight_layout()
plt.show()In the resulting plot, you can see that the polynomial fit captures the relationship between the response(Y) and explanatory variables(X) much better than the linear fit:
Next, we will compute the MSE and ![]() evaluation metrics:
evaluation metrics:
y_lin_pred = lr.predict(X)
y_quad_pred = pr.predict(X_quad)
print( 'Training MSE linear: %.3f, quadratic: %.3f' % ( mean_squared_error(y, y_lin_pred),
mean_squared_error(y, y_quad_pred)
) )
print( 'Train R^2 linear: %.3f, quadratic: %.3f' % ( r2_score(y, y_lin_pred),
r2_score(y, y_quad_pred)
) ) As you can see after executing the code, the MSE decreased from 570 (linear fit) to 61 (quadratic fit); also, the coefficient of determination决定系数 (![]() ) reflects a closer fit of the quadratic model (
) reflects a closer fit of the quadratic model (![]() = 0.982 ) as opposed to the linear fit (
= 0.982 ) as opposed to the linear fit (![]() = 0.832 ) in this particular toy problem.a high R2 value indicates that the model is a good fit for the data
= 0.832 ) in this particular toy problem.a high R2 value indicates that the model is a good fit for the data
Modeling nonlinear relationships in the Housing dataset
In the preceding subsection, you learned how to construct polynomial features to fit nonlinear relationships in a toy玩物/简单 problem; let's now take a look at a more concrete example and apply those concepts to the data in the Housing dataset. By executing the following code, we will model the relationship between house prices and LSTAT (percentage of lower status of the population) using second-degree (quadratic) and third-degree (cubic) polynomials and compare that to a linear fit:
df.head()
###########################################################################
class sklearn.preprocessing.PolynomialFeatures(degree=2, *, interaction_only=False, include_bias=True, order='C')Parameters
- degree : int, default=2
The degree of the polynomial features. - interaction_only : bool, default=False
If true, only interaction features are produced: features that are products of at mostdegreedistinct input features (so notx[1]**2,x[0]*x[2]**3, etc.). - include_bias : bool, default=True
If True (default), then include a bias column, the feature in which all polynomial powers are zero (i.e. a column of ones - acts as an intercept term in a linear model). - order : {‘C’, ‘F’}, default=’C’
Order of output array in the dense case. ‘F’ order is faster to compute, but may slow down subsequent estimators.
Consider the 2D array arr = np.arange(12).reshape(3,4). It looks like this:
In a computer memory this is stored as ![]()
This is also known as row-major order methods for storing multidimensional arrays in linear storage such as random access memory.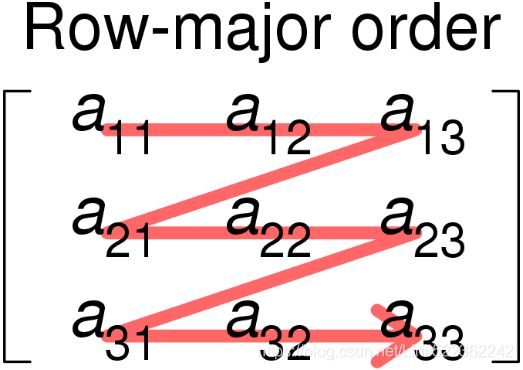 It is also called as
It is also called as C Contiguous layout (order ‘C’) because C arrays always start at zero and stored as contiguous block of memory. So the next memory address holds the next row value on that row.
For example, the array ![]()
The array A elements in C arrays are row major order as shown in this table https://en.wikipedia.org/wiki/Row-_and_column-major_order#/media/File:Row_and_column_major_order.svg
Fortran Contiguous or Column major
if you transpose the above array then it becomes Fortran contiguous layout i.e. order ‘F’
[ [a11, a21],
[a12, a22],
[a13, a23] ]
By default Fortran arrays start at 1 and also known as column-major order methods for storing multidimensional arrays in linear storage such as RAM
The array A elements in F contiguous arrays are column major order as shown in below table
The C contiguous memory layout, row-wise operations are usually faster than column-wise operations(A typical rowwise operation is to compute row means or row sums, for example to compute person sum scores for psychometric analyses.)
Similarly column wise operation is faster for F contiguous array
So we have understood what is conguous array and what is order difference between C and Fortran(F) and now we can explore numpy.ravel() and numpy.reshape()
numpy ravel explained
numpy.ravel(array, order=’C’)order = {‘C’,’F’, ‘A’, ‘K’}, optional and by default it is ‘C’
‘A’ means to read the elements in Fortran-like index order if array is Fortran contiguous in memory, C-like order otherwise
and ‘K’ means to read the elements in the order they occur in memory
x = np.array([[1, 2, 3], [4, 5, 6]])
xnp.ravel(x)np.ravel(x, order='F')numpy ravel with order A
x.Tnp.ravel(x.T)np.ravel(x.T,order='A')numpy.reshape()
numpy.reshape(a, newshape, order=’C’)order = {‘C’,’F’, ‘A’, ‘K’}, optional and by default it is ‘C’
x = np.array([[1, 2, 3], [4, 5, 6]])
x![]()
This a 2x3 array and we want to reshape it to 3x2
np.reshape(x,(3,2))![]()
np.reshape(np.ravel(x),(3,2))np.reshape(np.ravel(x),(3,-1)) The output is same here as previous one with shape (3,2) because when dimension -1 is given then it infers the value as 2 based on original shape of array
The output is same here as previous one with shape (3,2) because when dimension -1 is given then it infers the value as 2 based on original shape of array
###########################################################################
X = df[['LSTAT']].values
y = df['MEDV'].values
regr = LinearRegression()
# create quadratic features
quadratic = PolynomialFeatures( degree=2 )
cubic = PolynomialFeatures( degree=3 )
X_quad = quadratic.fit_transform(X)
X_cubic = cubic.fit_transform(X)
# fit features
X_new = np.arange( X.min(), X.max(), 1 )[:, np.newaxis] # 1D==>2D
regr = regr.fit(X,y)
linear_r2 = r2_score(y, regr.predict(X)) # get the training r2 score
y_new_lin_fit = regr.predict(X_new)
regr = regr.fit(X_quad, y)
quadratic_r2 = r2_score(y, regr.predict(X_quad)) # get the training r2 score
y_new_quad_fit = regr.predict( quadratic.fit_transform(X_new) )
regr = regr.fit(X_cubic, y)
cubic_r2 = r2_score(y, regr.predict(X_cubic)) # get the training r2 score
y_new_cubic_fit = regr.predict( cubic.fit_transform(X_new) )
#plot results
plt.figure(figsize=(10,8))
plt.scatter( X,y, label='Traning points', color='lightgray')
plt.plot( X_new, y_new_lin_fit,
label = 'Linear (d=1), $R^2=%.2f$' % linear_r2,
color = 'blue',
lw = 2,
linestyle = ':' )
plt.plot( X_new, y_new_quad_fit,
label = 'Quadratic (d=2), $R^2=%.2f$' % quadratic_r2,
color = 'red',
lw = 2,
ls = '-' )
plt.plot( X_new, y_new_cubic_fit,
label = 'Cubic (d=3), $R^2=%.2f$' % cubic_r2,
color = 'green',
lw = 2,
ls = '--' )
plt.xlabel( '% lower status of the population [LSTAT]' )
plt.ylabel( 'Price in $1000s [MEDV]' )
plt.legend( loc='upper right' )
plt.show()The resulting plot is as follows:
LSTAT : Percentage of lower status地位 of the population
##########################################################################
Why MEDV vs LSTAT since and
and ![]() the largest correlation with the LSTAT
the largest correlation with the LSTAT
variable (-0.74)
the correlation matrix(rescaled version of the covariance matrix, a correlation matrix to quantify量化 and summarize linear relationships between variables)
##########################################################################
As you can see, the cubic fit(with the highest R2 score) captures the relationship between house prices and LSTAT better than the linear and quadratic fit. However, you should be aware that adding more and more polynomial features increases the complexity of a model and therefore increases the chance of overfitting. Thus, in practice, it is always recommended to evaluate the performance of the model on a separate test dataset to estimate the generalization performance.
In addition, polynomial features are not always the best choice for modeling nonlinear relationships. For example, with some experience or intuition, just looking at the MEDV-LSTAT scatterplot may lead to the hypothesis that a log-transformation of the LSTAT feature variable and the square root of MEDV(Median value of owner-occupied homes in $1000s) may project the data onto a linear feature space suitable for a linear regression fit. For instance, my perception is that this relationship between the two variables looks quite similar to an exponential function:![]()
 Note (e^-1)^x and 0<(e^-1) <1
Note (e^-1)^x and 0<(e^-1) <1
Since the natural logarithm of an exponential function is a straight line, I assume that such a log-transformation can be usefully applied here:![]()
Let's test this hypothesis by executing the following code:
# transform features
X_log = np.log(X) # since the hypothesis: y=e^(-X), then log(e^-X)=-X ==> a straight line
y_sqrt = np.sqrt(y)
# fit features
X_new_log = np.arange( X_log.min()-1, X_log.max()+1, 1)[:, np.newaxis]
regr = regr.fit( X_log, y_sqrt )
linear_r2 = r2_score( y_sqrt, regr.predict(X_log) )
y_new_lin_fit = regr.predict( X_new_log )
# plot results
plt.scatter( X_log, y_sqrt, label='Training poits', color='lightgray' )
plt.plot( X_new_log, y_new_lin_fit,
label = 'Linear (d=1), $R^2=%.2f$' % linear_r2,
color = 'blue',
lw=2 )
plt.xlabel( 'log(% lower status of the population [LSTAT])' )
plt.ylabel( '$\sqrt{Price \; in \; \$1000s \; [MEDV]}$' ) # OR # plt.ylabel( '$\sqrt{Price\;in\;\$1000s\;[MEDV]}$' )
plt.legend( loc='lower left' )
plt.tight_layout()
plt.show() After transforming the explanatory onto the log space and taking the square root of the target variables, we were able to capture the relationship between the two variables with a linear regression line that seems to fit the data better (2 = 0.69 ) than any of the previous polynomial feature transformations:
Dealing with nonlinear relationships using random forests
In this section, we are going to take a look at random forest regression, which is conceptually different from the previous regression models in this chapter. A random forest, which is an ensemble of multiple decision trees, can be understood as the sum of piecewise linear functions, in contrast to the global linear and polynomial regression models that we discussed previously. In other words, via the decision tree algorithm, we subdivide the input space into smaller regions that become more manageable.
Decision tree regression
An advantage of the decision tree algorithm is that it does not require any transformation of the features if we are dealing with nonlinear data, because decision trees analyze one feature at a time, rather than taking weighted combinations into account. (Likewise, normalizing or standardizing features is not required for decision trees.) You will remember from Cp3 ML Classifiers_2_support vector_Maximum margin_soft margin_C~slack_kernel_Gini_pydot+_Information Gain https://blog.csdn.net/Linli522362242/article/details/107755405, that we grow a decision tree by iteratively splitting its nodes until the leaves are pure or a stopping criterion is satisfied. When we used decision trees for classification, we defined entropy as a measure of impurity to determine which feature split maximizes the information gain (IG) , which can be defined as follows for a binary split: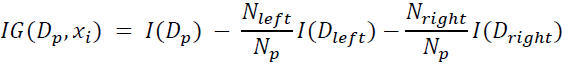 (信息增益=父节点data set的总不纯度(含有有用的信息)-权重*左节点data set的不纯度-权重*右节点的data set不纯度,每次分割都能获得信息增益)
(信息增益=父节点data set的总不纯度(含有有用的信息)-权重*左节点data set的不纯度-权重*右节点的data set不纯度,每次分割都能获得信息增益)
Here, ![]() is the subset of training examples at the parent node,
is the subset of training examples at the parent node, ![]() is the feature to perform the split,
is the feature to perform the split, ![]() is the total number of samples at the parent node, I is the impurity function, , and
is the total number of samples at the parent node, I is the impurity function, , and ![]() and
and ![]() are the subsets of training examples at the left and right child nodes after the split.
are the subsets of training examples at the left and right child nodes after the split. ![]() is the number of samples in the jth child node (Here j=left or right). Remember that our goal is to find the feature split that maximizes the information gain; in other words, we want to find the feature split that reduces the impurities in the child nodes most(找到特征划分使得的子节点中信息不纯度最低(或者最大减少子节点的不纯度)). In Cp3 ML Classifiers_2_support vector_Maximum margin_soft margin_C~slack_kernel_Gini_pydot+_Information Gain https://blog.csdn.net/Linli522362242/article/details/107755405, we discussed
is the number of samples in the jth child node (Here j=left or right). Remember that our goal is to find the feature split that maximizes the information gain; in other words, we want to find the feature split that reduces the impurities in the child nodes most(找到特征划分使得的子节点中信息不纯度最低(或者最大减少子节点的不纯度)). In Cp3 ML Classifiers_2_support vector_Maximum margin_soft margin_C~slack_kernel_Gini_pydot+_Information Gain https://blog.csdn.net/Linli522362242/article/details/107755405, we discussed
Gini impurity (Gini index, as a measure of node purity—a small value indicates that a node contains predominantly显著地 observations(or samples) from a single class(c=1);  0 ≤
0 ≤ ![]() ≤ 1, OR
≤ 1, OR ![]() 0 ≤ ˆpmk ≤ 1;
0 ≤ ˆpmk ≤ 1;
########################################06_Decision Trees_01_graphviz_Gini_Entropy_Decision Tree_CART_prune_Regression_Classification_Treehttps://blog.csdn.net/Linli522362242/article/details/104542381
To compute Gini impurity for a set of items with ![]() classes, suppose
classes, suppose ![]() , and let
, and let ![]() be the fraction of items labeled with class
be the fraction of items labeled with class ![]() in the set. (
in the set. (![]() is the proportion of class i in the set)
is the proportion of class i in the set) 
![]()
![]() : the sum of all probabilities of all classes(J) is equal to 1.
: the sum of all probabilities of all classes(J) is equal to 1.
example: 
![]() and 1 ==
and 1 == ![]() +
+ ![]() +
+ ![]()
########################################https://blog.csdn.net/Linli522362242/article/details/104542381
eg, K=2, means the mth node is relatively pure, p=0.9, p *q= 0.9*0.1== 0.09, Gini index at mth node=0.9*0.1+0.1*0.9=0.18; the Gini impurity is maximal if the classes are perfectly mixed(distributed uniformly), for example, in a binary class setting ( ![]() ):
): 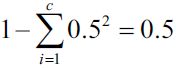
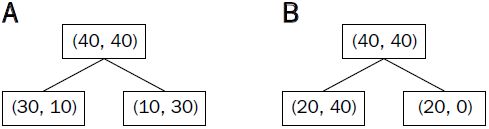
We start with a dataset ![]() at the parent node
at the parent node ![]() that consists of 40 samples from class 1 and 40 samples from class 2 that we split into two datasets
that consists of 40 samples from class 1 and 40 samples from class 2 that we split into two datasets ![]() and
and ![]() , respectively.
, respectively.
However, the Gini impurity would favor the split in scenario B(
would favor the split in scenario B( ![]() ) over scenario A(
) over scenario A(![]() ), which is indeed more pure:
), which is indeed more pure: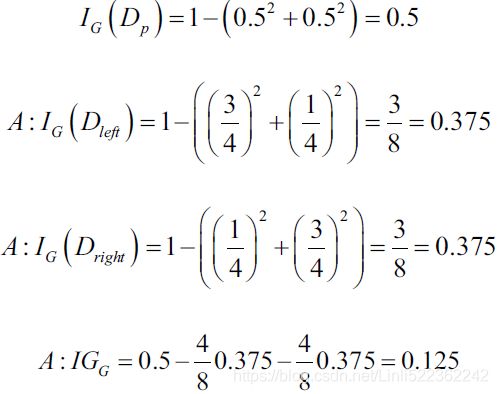
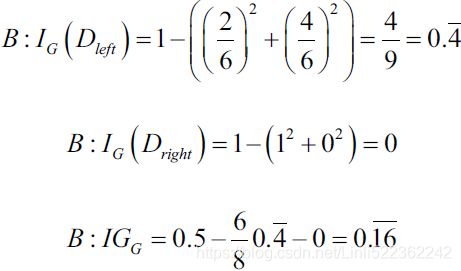
)
and entropy (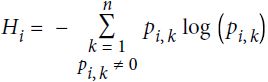 OR
OR![]() 0 ≤
0 ≤ ![]() ≤ 1, Here,
≤ 1, Here, ![]() is the proportion of the samples that belongs to class i for a particular node t. The entropy is therefore 0 if all samples at a node belong to the same class, and the entropy is maximal if we have a uniform class distribution. For example, in a binary class setting, the entropy is 0(= -(1*0 + 0*
is the proportion of the samples that belongs to class i for a particular node t. The entropy is therefore 0 if all samples at a node belong to the same class, and the entropy is maximal if we have a uniform class distribution. For example, in a binary class setting, the entropy is 0(= -(1*0 + 0*![]() (0)) ) if p (i =1| t ) =1 or p(i = 0 | t ) = 0. If the classes are distributed uniformly with p (i =1| t ) = 0.5 and p(i = 0 | t ) = 0.5, the entropy is 1(= - (0.5*-1 + 0.5*-1) ) . Therefore, we can say that the entropy criterion attempts to maximize the mutual information in the tree.
(0)) ) if p (i =1| t ) =1 or p(i = 0 | t ) = 0. If the classes are distributed uniformly with p (i =1| t ) = 0.5 and p(i = 0 | t ) = 0.5, the entropy is 1(= - (0.5*-1 + 0.5*-1) ) . Therefore, we can say that the entropy criterion attempts to maximize the mutual information in the tree.
##########################################https://blog.csdn.net/Linli522362242/article/details/104542381
We start with a dataset ![]() at the parent node
at the parent node ![]() that consists of 40 samples from class 1 and 40 samples from class 2 that we split into two datasets
that consists of 40 samples from class 1 and 40 samples from class 2 that we split into two datasets ![]() and
and ![]() , respectively.
, respectively.
the entropy criterion ![]() would favor scenario B(
would favor scenario B( ![]() ) over scenario (
) over scenario (![]() ) :
) :

Gini impurity is slightly faster to compute, so it is a good default. However, when they differ, Gini impurity tends to isolate the most frequent class in its own branch of the tree, while entropy tends to produce slightly more balanced trees.
(more times of spliting a dataset, larger size to the tree, more number of leaf nodes(and each leaf may just contain fewer data points or samples) and the model become more complexity, but minimizes residual errors )
##########################################https://blog.csdn.net/Linli522362242/article/details/104542381
)
as measures of impurity, which are both useful criteria for classification. To use a decision tree for regression, however, we need an impurity metric that is suitable for continuous variables, so we define the impurity measure of a node, t, as the MSE instead:
Here, ![]() is the number of training samples at node t ,
is the number of training samples at node t , ![]() is the training subset at node t ,
is the training subset at node t , ![]() is the true target value, and
is the true target value, and ![]() is the predicted target value (sample mean):
is the predicted target value (sample mean):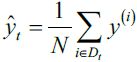
############################
The pruning is based on a criterion that balances residual error against a measure of model complexity. If we denote the starting tree for pruning by ![]() , then we define
, then we define ![]() to be a subtree of
to be a subtree of ![]() if it can be obtained by pruning nodes from
if it can be obtained by pruning nodes from ![]() (in other words, by collapsing折叠 internal nodes by combining the corresponding regions即通过合并对应区域来收缩内部结点). Suppose the leaf nodes are indexed by τ = 1, . . . , |T|, with leaf node τ representing a region
(in other words, by collapsing折叠 internal nodes by combining the corresponding regions即通过合并对应区域来收缩内部结点). Suppose the leaf nodes are indexed by τ = 1, . . . , |T|, with leaf node τ representing a region ![]() of input space having
of input space having ![]() data points(samples), and |T| denoting the total number of leaf nodes. The optimal prediction for region
data points(samples), and |T| denoting the total number of leaf nodes. The optimal prediction for region ![]() is then given by 那么区域
is then given by 那么区域![]() 给出的最优的预测为
给出的最优的预测为 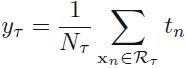
and the corresponding contribution to the residual sum-of-squares is then 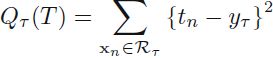
The pruning criterion is then given by ![]() (minimizing)
(minimizing)
The regularization parameter λ determines the trade-off between the overall residual sum-of-squares error and the complexity of the model as measured by the number |T| of leaf nodes, and its value is chosen by cross-validation.
########################################################06_Decision Trees_01_graphviz_Gini_Entropy_Decision Tree_CART_prune_Regression_Classification_Treehttps://blog.csdn.net/Linli522362242/article/details/104542381
In the context of decision tree regression, the MSE is often also referred to as within-node variance, which is why the splitting criterion is also better known as variance reduction. To see what the line fit of a decision tree looks like, let's use the DecisionTreeRegressor implemented in scikit-learn to model the nonlinear relationship between the MEDV and LSTAT variables:
from sklearn.tree import DecisionTreeRegressor
X = df[['LSTAT']].values
y = df['MEDV'].values
tree = DecisionTreeRegressor( max_depth=3 )
tree.fit(X,y)![]()
def lin_regplot(X,y, model):
plt.scatter( X,y, c='steelblue', edgecolor='white', s=70 )
plt.plot(X, model.predict(X), color='black', lw=2 )
return # or # return None
sort_idx = X.flatten().argsort() # return index after sorting X.flatten()
lin_regplot(X[sort_idx], y[sort_idx], tree)
plt.xlabel('% lower status of the population [LSTAT]')
plt.ylabel('Price in $1000s [MEDV]')
plt.show() As we can see from the resulting plot, the decision tree captures the general trend in the data. However, a limitation of this model is that it does not capture the continuity and differentiability of the desired prediction. In addition, we need to be careful about choosing an appropriate value for the depth of the tree to not overfit or underfit the data; here, a depth of 3 seems to be a good choice: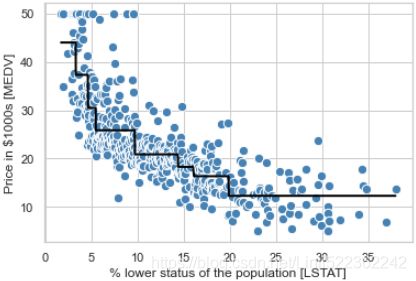 2^3=8
2^3=8
In the next section, we will take a look at a more robust way for fitting regression trees: random forests.
#########################################################
class sklearn.tree.DecisionTreeRegressor(*, criterion='mse', splitter='best', max_depth=None, min_samples_split=2,
min_samples_leaf=1, min_weight_fraction_leaf=0.0, max_features=None,
random_state=None, max_leaf_nodes=None, min_impurity_decrease=0.0,
min_impurity_split=None, ccp_alpha=0.0)criterion : {“mse”, “friedman_mse”, “mae”, “poisson”}, default=”mse”
The function to measure the quality of a split. Supported criteria are “mse” for the mean squared error, which is equal to variance reduction as feature selection criterion and minimizes the L2 loss using the mean of each terminal node, “friedman_mse”, which uses mean squared error with Friedman’s improvement score for potential splits, “mae” for the mean absolute error, which minimizes the L1 loss using the median of each terminal node, and “poisson” which uses reduction in Poisson deviance to find splits.
as feature selection criterion and minimizes the L2 loss using the mean of each terminal node, “friedman_mse”, which uses mean squared error with Friedman’s improvement score for potential splits, “mae” for the mean absolute error, which minimizes the L1 loss using the median of each terminal node, and “poisson” which uses reduction in Poisson deviance to find splits.
max_depth : int, default=None
The maximum depth of the tree. If None, then nodes are expanded until all leaves are pure or until all leaves contain less than min_samples_split samples.
min_impurity_decrease : float, default=0.0
A node will be split if this split induces a decrease of the impurity greater than or equal to this value.
The weighted impurity decrease equation is the following:
N_t / N * (impurity - N_t_R / N_t * right_impurity
- N_t_L / N_t * left_impurity) where N is the total number of samples, N_t is the number of samples at the current node, N_t_L is the number of samples in the left child, and N_t_R is the number of samples in the right child.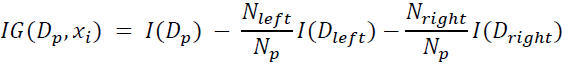
#########################################################
Random forest regression
As we discussed in cp3 A Tour of ML Classifiers_3_ random forests_knn https://blog.csdn.net/Linli522362242/article/details/107843678, the random forest algorithm is an ensemble technique that combines multiple decision trees. A random forest usually has a better generalization performance than an individual decision tree due to randomness that helps to decrease the model variance. Other advantages of random forests are that they are less sensitive to outliers in the dataset and don't require much parameter tuning. The only parameter in random forests that we typically need to experiment with is the number of trees in the ensemble. The basic random forests algorithm for regression is almost identical to the random forest algorithm for classification that
we discussed in Cp3. The only difference is that we use the MSE criterion to grow the individual decision trees, and the predicted target variable is calculated as the average prediction over all decision trees.
Now, let's use all the features in the Housing Dataset to fit a random forest regression model on 60 percent of the samples and evaluate its performance on the remaining 40 percent. The code is as follows:
from sklearn.ensemble import RandomForestRegressor
forest = RandomForestRegressor( n_estimators=1000,
criterion='mse',
random_state=1,
n_jobs=-1)
forest.fit(X_train, y_train)
y_train_pred = forest.predict(X_train)
y_test_pred = forest.predict(X_test)
print( 'MSE train: %.3f, test: %.3f' % ( mean_squared_error(y_train, y_train_pred),
mean_squared_error(y_test, y_test_pred)
) )
print( 'R^2 train: %.3f, test: %.3f' % ( r2_score(y_train, y_train_pred),
r2_score(y_test, y_test_pred)
) )![]()
Unfortunately, we see that the random forest tends to overfit the training data. However, it's still able to explain the relationship between the target and explanatory variables relatively well ( R2 = 0.871 on the test dataset).
Lastly, let's also take a look at the residuals of the prediction:
plt.scatter( y_train_pred,
y_train_pred - y_train,
c='steelblue',
edgecolors='white',
marker='o',
s=35,
alpha=0.9,
label='Training data' )
plt.scatter( y_test_pred,
y_test_pred - y_test,
c='limegreen',
edgecolors='white',
marker='s',
s=35,
alpha=0.9,
label='Test data')
plt.xlabel('Predicted values')
plt.ylabel('Residuals')
plt.legend(loc='upper left')
plt.hlines(y=0, xmin=-10, xmax=50, lw=2, color='black')
plt.xlim([-10, 50])
plt.tight_layout()
plt.show()As it was already summarized by the R2 coefficient, we can see that the model fits the training data better than the test data, as indicated by the outliers in the y axis direction. Also, the distribution of the residuals does not seem to be completely random around the zero center point, indicating that the model is not able to capture all the exploratory information. However, the residual plot indicates a large improvement over the residual plot of the linear model that we plotted earlier in this chapter:
Summary
At the beginning of this chapter, you learned about using simple linear regression analysis to model the relationship between a single explanatory variable and a continuous response variable. We then discussed a useful explanatory data analysis technique to look at patterns and anomalies in data, which is an important first step in predictive modeling tasks.
We built our first model by implementing linear regression using a gradient-based optimization approach. We then saw how to utilize scikit-learn's linear models for regression and also implement a robust regression technique (RANSAC) as an approach for dealing with outliers. To assess the predictive performance of regression models, we computed the mean sum of squared errors and the related R2 metric. Furthermore, we also discussed a useful graphical approach to diagnose the problems of regression models: the residual plot.
After we discussed how regularization can be applied to regression models to reduce the model complexity and avoid overfitting, we also introduced several approaches to model nonlinear relationships, including polynomial feature transformation and random forest regressors.
We discussed supervised learning, classification, and regression analysis, in great detail throughout the previous chapters. In the next chapter, we are going to discuss another interesting subfield of machine learning: unsupervised learning. In the next chapter, you will learn how to use cluster analysis for finding hidden structures in data in the absence of target variables.