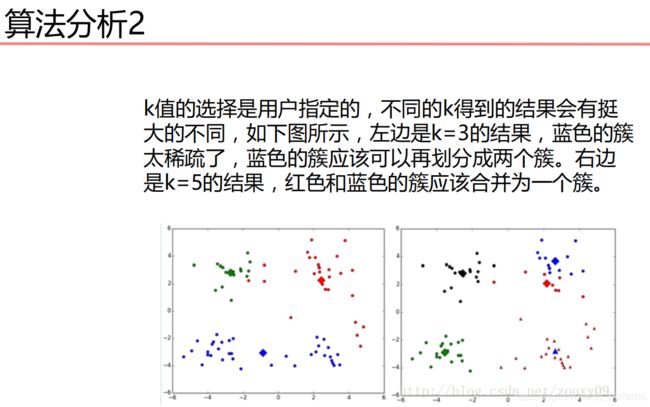
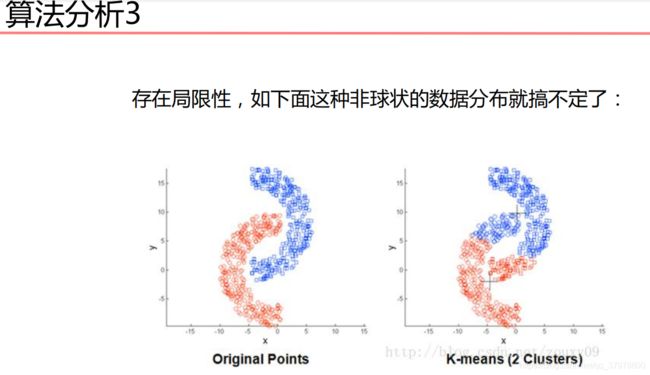
机器学习基础-聚类算法-15
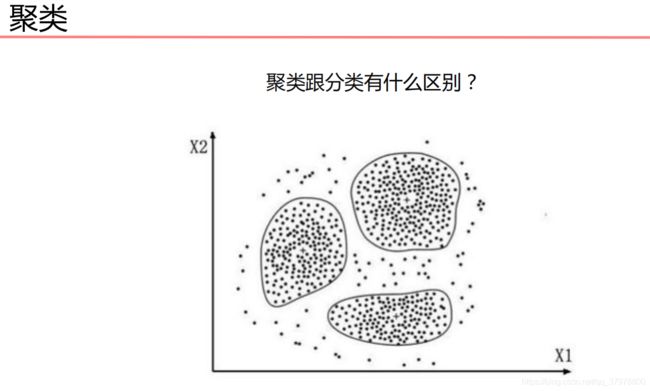
聚类算法
K-MEANS
python实现K-MEANS
import numpy as np
import matplotlib.pyplot as plt
# 载入数据
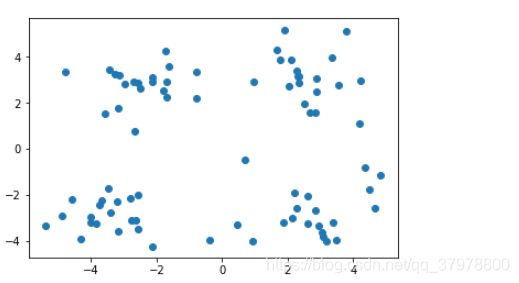
data = np.genfromtxt("kmeans.txt", delimiter=" ")
plt.scatter(data[:,0],data[:,1])
plt.show()
# 计算距离
def euclDistance(vector1, vector2):
return np.sqrt(sum((vector2 - vector1)**2))
# 初始化质心
def initCentroids(data, k):
numSamples, dim = data.shape
# k个质心,列数跟样本的列数一样
centroids = np.zeros((k, dim))
# 随机选出k个质心
for i in range(k):
# 随机选取一个样本的索引
index = int(np.random.uniform(0, numSamples))
# 作为初始化的质心
centroids[i, :] = data[index, :]
return centroids
# 传入数据集和k的值
def kmeans(data, k):
# 计算样本个数
numSamples = data.shape[0]
# 样本的属性,第一列保存该样本属于哪个簇,第二列保存该样本跟它所属簇的误差
clusterData = np.array(np.zeros((numSamples, 2)))
# 决定质心是否要改变的变量
clusterChanged = True
# 初始化质心
centroids = initCentroids(data, k)
while clusterChanged:
clusterChanged = False
# 循环每一个样本
for i in range(numSamples):
# 最小距离
minDist = 100000.0
# 定义样本所属的簇
minIndex = 0
# 循环计算每一个质心与该样本的距离
for j in range(k):
# 循环每一个质心和样本,计算距离
distance = euclDistance(centroids[j, :], data[i, :])
# 如果计算的距离小于最小距离,则更新最小距离
if distance < minDist:
minDist = distance
# 更新最小距离
clusterData[i, 1] = minDist
# 更新样本所属的簇
minIndex = j
# 如果样本的所属的簇发生了变化
if clusterData[i, 0] != minIndex:
# 质心要重新计算
clusterChanged = True
# 更新样本的簇
clusterData[i, 0] = minIndex
# 更新质心
for j in range(k):
# 获取第j个簇所有的样本所在的索引
cluster_index = np.nonzero(clusterData[:, 0] == j)
# 第j个簇所有的样本点
pointsInCluster = data[cluster_index]
# 计算质心
centroids[j, :] = np.mean(pointsInCluster, axis = 0)
# showCluster(data, k, centroids, clusterData)
return centroids, clusterData
# 显示结果
def showCluster(data, k, centroids, clusterData):
numSamples, dim = data.shape
if dim != 2:
print("dimension of your data is not 2!")
return 1
# 用不同颜色形状来表示各个类别
mark = ['or', 'ob', 'og', 'ok', '^r', '+r', 'sr', 'dr', ', 'pr']
if k > len(mark):
print("Your k is too large!")
return 1
# 画样本点
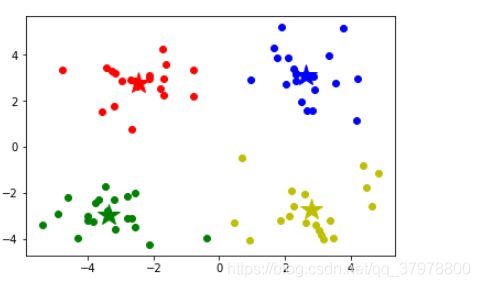
for i in range(numSamples):
markIndex = int(clusterData[i, 0])
plt.plot(data[i, 0], data[i, 1], mark[markIndex])
# 用不同颜色形状来表示各个类别
mark = ['*r', '*b', '*g', '*k', '^b', '+b', 'sb', 'db', ', 'pb']
# 画质心点
for i in range(k):
plt.plot(centroids[i, 0], centroids[i, 1], mark[i], markersize = 20)
plt.show()
# 设置k值
k = 4
# centroids 簇的中心点
# cluster Data样本的属性,第一列保存该样本属于哪个簇,第二列保存该样本跟它所属簇的误差
centroids, clusterData = kmeans(data, k)
if np.isnan(centroids).any():
print('Error')
else:
print('cluster complete!')
# 显示结果
showCluster(data, k, centroids, clusterData)
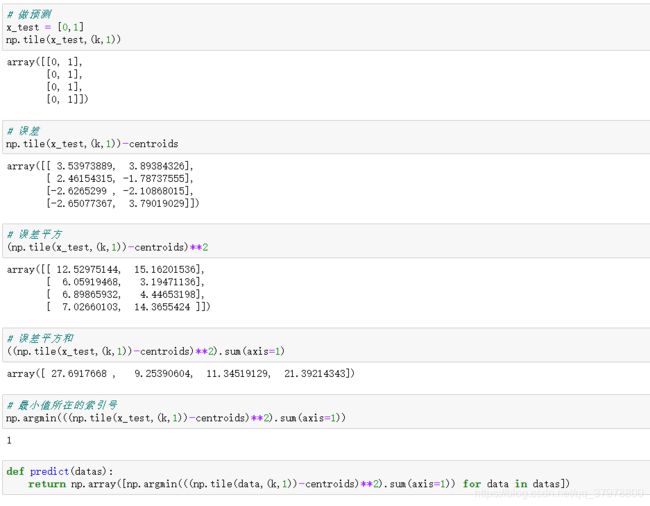
def predict(datas):
return np.array([np.argmin(((np.tile(data,(k,1))-centroids)**2).sum(axis=1)) for data in datas])
画出簇的作用区域
# 获取数据值所在的范围
x_min, x_max = data[:, 0].min() - 1, data[:, 0].max() + 1
y_min, y_max = data[:, 1].min() - 1, data[:, 1].max() + 1
# 生成网格矩阵
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.02),
np.arange(y_min, y_max, 0.02))
z = predict(np.c_[xx.ravel(), yy.ravel()])# ravel与flatten类似,多维数据转一维。flatten不会改变原始数据,ravel会改变原始数据
z = z.reshape(xx.shape)
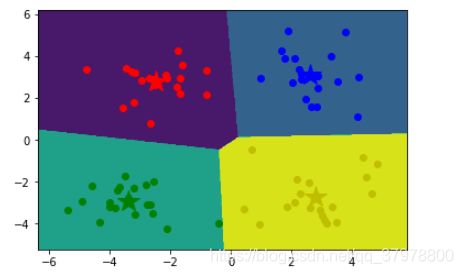
# 等高线图
cs = plt.contourf(xx, yy, z)
# 显示结果
showCluster(data, k, centroids, clusterData)
sklearn-K-MEANS
from sklearn.cluster import KMeans
import numpy as np
import matplotlib.pyplot as plt
# 载入数据
data = np.genfromtxt("kmeans.txt", delimiter=" ")
# 设置k值
k = 4
# 训练模型
model = KMeans(n_clusters=k)
model.fit(data)
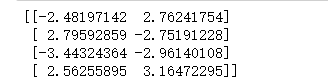
# 分类中心点坐标
centers = model.cluster_centers_
print(centers)
# 预测结果
result = model.predict(data)
print(result)
model.labels_
# 画出各个数据点,用不同颜色表示分类
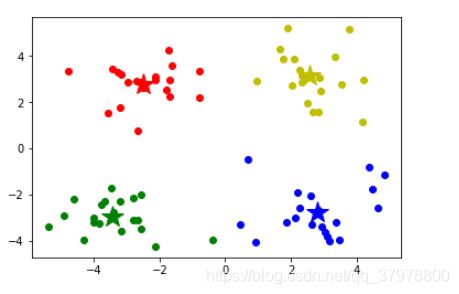
mark = ['or', 'ob', 'og', 'oy']
for i,d in enumerate(data):
plt.plot(d[0], d[1], mark[result[i]])
# 画出各个分类的中心点
mark = ['*r', '*b', '*g', '*y']
for i,center in enumerate(centers):
plt.plot(center[0],center[1], mark[i], markersize=20)
plt.show()
# 获取数据值所在的范围
x_min, x_max = data[:, 0].min() - 1, data[:, 0].max() + 1
y_min, y_max = data[:, 1].min() - 1, data[:, 1].max() + 1
# 生成网格矩阵
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.02),
np.arange(y_min, y_max, 0.02))
z = model.predict(np.c_[xx.ravel(), yy.ravel()])# ravel与flatten类似,多维数据转一维。flatten不会改变原始数据,ravel会改变原始数据
z = z.reshape(xx.shape)
# 等高线图
cs = plt.contourf(xx, yy, z)
# 显示结果
# 画出各个数据点,用不同颜色表示分类
mark = ['or', 'ob', 'og', 'oy']
for i,d in enumerate(data):
plt.plot(d[0], d[1], mark[result[i]])
# 画出各个分类的中心点
mark = ['*r', '*b', '*g', '*y']
for i,center in enumerate(centers):
plt.plot(center[0],center[1], mark[i], markersize=20)
plt.show()
Mini Batch K-Means
from sklearn.cluster import MiniBatchKMeans
import numpy as np
import matplotlib.pyplot as plt
# 载入数据
data = np.genfromtxt("kmeans.txt", delimiter=" ")
# 设置k值
k = 4
# 训练模型
model = MiniBatchKMeans(n_clusters=k)
model.fit(data)
# 分类中心点坐标
centers = model.cluster_centers_
print(centers)
# 预测结果
result = model.predict(data)
print(result)
# 画出各个数据点,用不同颜色表示分类
mark = ['or', 'ob', 'og', 'oy']
for i,d in enumerate(data):
plt.plot(d[0], d[1], mark[result[i]])
# 画出各个分类的中心点
mark = ['*r', '*b', '*g', '*y']
for i,center in enumerate(centers):
plt.plot(center[0],center[1], mark[i], markersize=20)
plt.show()
# 获取数据值所在的范围
x_min, x_max = data[:, 0].min() - 1, data[:, 0].max() + 1
y_min, y_max = data[:, 1].min() - 1, data[:, 1].max() + 1
# 生成网格矩阵
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.02),
np.arange(y_min, y_max, 0.02))
z = model.predict(np.c_[xx.ravel(), yy.ravel()])# ravel与flatten类似,多维数据转一维。flatten不会改变原始数据,ravel会改变原始数据
z = z.reshape(xx.shape)
# 等高线图
cs = plt.contourf(xx, yy, z)
# 显示结果
# 画出各个数据点,用不同颜色表示分类
mark = ['or', 'ob', 'og', 'oy']
for i,d in enumerate(data):
plt.plot(d[0], d[1], mark[result[i]])
# 画出各个分类的中心点
mark = ['*r', '*b', '*g', '*y']
for i,center in enumerate(centers):
plt.plot(center[0],center[1], mark[i], markersize=20)
plt.show()
python实现K-MEANS优化1
import numpy as np
import matplotlib.pyplot as plt
# 载入数据
data = np.genfromtxt("kmeans.txt", delimiter=" ")
训练模型
# 计算距离
def euclDistance(vector1, vector2):
return np.sqrt(sum((vector2 - vector1)**2))
# 初始化质心
def initCentroids(data, k):
numSamples, dim = data.shape
# k个质心,列数跟样本的列数一样
centroids = np.zeros((k, dim))
# 随机选出k个质心
for i in range(k):
# 随机选取一个样本的索引
index = int(np.random.uniform(0, numSamples))
# 作为初始化的质心
centroids[i, :] = data[index, :]
return centroids
# 传入数据集和k的值
def kmeans(data, k):
# 计算样本个数
numSamples = data.shape[0]
# 样本的属性,第一列保存该样本属于哪个簇,第二列保存该样本跟它所属簇的误差
clusterData = np.array(np.zeros((numSamples, 2)))
# 决定质心是否要改变的变量
clusterChanged = True
# 初始化质心
centroids = initCentroids(data, k)
while clusterChanged:
clusterChanged = False
# 循环每一个样本
for i in range(numSamples):
# 最小距离
minDist = 100000.0
# 定义样本所属的簇
minIndex = 0
# 循环计算每一个质心与该样本的距离
for j in range(k):
# 循环每一个质心和样本,计算距离
distance = euclDistance(centroids[j, :], data[i, :])
# 如果计算的距离小于最小距离,则更新最小距离
if distance < minDist:
minDist = distance
# 更新样本所属的簇
minIndex = j
# 更新最小距离
clusterData[i, 1] = distance
# 如果样本的所属的簇发生了变化
if clusterData[i, 0] != minIndex:
# 质心要重新计算
clusterChanged = True
# 更新样本的簇
clusterData[i, 0] = minIndex
# 更新质心
for j in range(k):
# 获取第j个簇所有的样本所在的索引
cluster_index = np.nonzero(clusterData[:, 0] == j)
# 第j个簇所有的样本点
pointsInCluster = data[cluster_index]
# 计算质心
centroids[j, :] = np.mean(pointsInCluster, axis = 0)
# showCluster(data, k, centroids, clusterData)
return centroids, clusterData
# 显示结果
def showCluster(data, k, centroids, clusterData):
numSamples, dim = data.shape
if dim != 2:
print("dimension of your data is not 2!")
return 1
# 用不同颜色形状来表示各个类别
mark = ['or', 'ob', 'og', 'ok', '^r', '+r', 'sr', 'dr', ', 'pr']
if k > len(mark):
print("Your k is too large!")
return 1
# 画样本点
for i in range(numSamples):
markIndex = int(clusterData[i, 0])
plt.plot(data[i, 0], data[i, 1], mark[markIndex])
# 用不同颜色形状来表示各个类别
mark = ['*r', '*b', '*g', '*k', '^b', '+b', 'sb', 'db', ', 'pb']
# 画质心点
for i in range(k):
plt.plot(centroids[i, 0], centroids[i, 1], mark[i], markersize = 20)
plt.show()
list_lost = []
for k in range(2,10):
min_loss = 10000
min_loss_centroids = np.array([])
min_loss_clusterData = np.array([])
for i in range(50):
# centroids 簇的中心点
# cluster Data样本的属性,第一列保存该样本属于哪个簇,第二列保存该样本跟它所属簇的误差
centroids, clusterData = kmeans(data, k)
loss = sum(clusterData[:,1])/data.shape[0]
if loss < min_loss:
min_loss = loss
min_loss_centroids = centroids
min_loss_clusterData = clusterData
list_lost.append(min_loss)
# print('loss',min_loss)
# print('cluster complete!')
# centroids = min_loss_centroids
# clusterData = min_loss_clusterData
# 显示结果
# showCluster(data, k, centroids, clusterData)
plt.plot(range(2,10),list_lost)
plt.xlabel('k')
plt.ylabel('loss')
plt.show()
# 获取数据值所在的范围
x_min, x_max = data[:, 0].min() - 1, data[:, 0].max() + 1
y_min, y_max = data[:, 1].min() - 1, data[:, 1].max() + 1
# 生成网格矩阵
xx, yy = np.meshgrid(np.arange(x_min, x_max, 0.02),
np.arange(y_min, y_max, 0.02))
z = predict(np.c_[xx.ravel(), yy.ravel()])# ravel与flatten类似,多维数据转一维。flatten不会改变原始数据,ravel会改变原始数据
z = z.reshape(xx.shape)
# 等高线图
cs = plt.contourf(xx, yy, z)
# 显示结果
showCluster(data, k, centroids, clusterData)
K-MEANS代价函数应用
import numpy as np
import matplotlib.pyplot as plt
# 载入数据
data = np.genfromtxt("kmeans.txt", delimiter=" ")
训练模型
# 计算距离
def euclDistance(vector1, vector2):
return np.sqrt(sum((vector2 - vector1)**2))
# 初始化质心
def initCentroids(data, k):
numSamples, dim = data.shape
# k个质心,列数跟样本的列数一样
centroids = np.zeros((k, dim))
# 随机选出k个质心
for i in range(k):
# 随机选取一个样本的索引
index = int(np.random.uniform(0, numSamples))
# 作为初始化的质心
centroids[i, :] = data[index, :]
return centroids
# 传入数据集和k的值
def kmeans(data, k):
# 计算样本个数
numSamples = data.shape[0]
# 样本的属性,第一列保存该样本属于哪个簇,第二列保存该样本跟它所属簇的误差
clusterData = np.array(np.zeros((numSamples, 2)))
# 决定质心是否要改变的变量
clusterChanged = True
# 初始化质心
centroids = initCentroids(data, k)
while clusterChanged:
clusterChanged = False
# 循环每一个样本
for i in range(numSamples):
# 最小距离
minDist = 100000.0
# 定义样本所属的簇
minIndex = 0
# 循环计算每一个质心与该样本的距离
for j in range(k):
# 循环每一个质心和样本,计算距离
distance = euclDistance(centroids[j, :], data[i, :])
# 如果计算的距离小于最小距离,则更新最小距离
if distance < minDist:
minDist = distance
# 更新样本所属的簇
minIndex = j
# 更新最小距离
clusterData[i, 1] = distance
# 如果样本的所属的簇发生了变化
if clusterData[i, 0] != minIndex:
# 质心要重新计算
clusterChanged = True
# 更新样本的簇
clusterData[i, 0] = minIndex
# 更新质心
for j in range(k):
# 获取第j个簇所有的样本所在的索引
cluster_index = np.nonzero(clusterData[:, 0] == j)
# 第j个簇所有的样本点
pointsInCluster = data[cluster_index]
# 计算质心
centroids[j, :] = np.mean(pointsInCluster, axis = 0)
# showCluster(data, k, centroids, clusterData)
return centroids, clusterData
# 显示结果
def showCluster(data, k, centroids, clusterData):
numSamples, dim = data.shape
if dim != 2:
print("dimension of your data is not 2!")
return 1
# 用不同颜色形状来表示各个类别
mark = ['or', 'ob', 'og', 'ok', '^r', '+r', 'sr', 'dr', ', 'pr']
if k > len(mark):
print("Your k is too large!")
return 1
# 画样本点
for i in range(numSamples):
markIndex = int(clusterData[i, 0])
plt.plot(data[i, 0], data[i, 1], mark[markIndex])
# 用不同颜色形状来表示各个类别
mark = ['*r', '*b', '*g', '*k', '^b', '+b', 'sb', 'db', ', 'pb']
# 画质心点
for i in range(k):
plt.plot(centroids[i, 0], centroids[i, 1], mark[i], markersize = 20)
plt.show()
# 设置k值
k = 4
min_loss = 10000
min_loss_centroids = np.array([])
min_loss_clusterData = np.array([])
for i in range(50):
# centroids 簇的中心点
# cluster Data样本的属性,第一列保存该样本属于哪个簇,第二列保存该样本跟它所属簇的误差
centroids, clusterData = kmeans(data, k)
loss = sum(clusterData[:,1])/data.shape[0]
if loss < min_loss:
min_loss = loss
min_loss_centroids = centroids
min_loss_clusterData = clusterData
# print('loss',min_loss)
print('cluster complete!')
centroids = min_loss_centroids
clusterData = min_loss_clusterData
# 显示结果
showCluster(data, k, centroids, clusterData)
DBSCAN
from sklearn.cluster import DBSCAN
import numpy as np
import matplotlib.pyplot as plt
# 载入数据
data = np.genfromtxt("kmeans.txt", delimiter=" ")
# 训练模型
# eps距离阈值,min_samples核心对象在eps领域的样本数阈值
model = DBSCAN(eps=1.5, min_samples=4)
model.fit(data)
result = model.fit_predict(data)
result
# 画出各个数据点,用不同颜色表示分类
mark = ['or', 'ob', 'og', 'oy', 'ok', 'om']
for i,d in enumerate(data):
plt.plot(d[0], d[1], mark[result[i]])
plt.show()
import numpy as np
import matplotlib.pyplot as plt
from sklearn import datasets
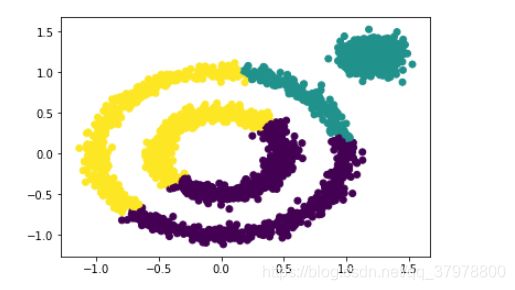
x1, y1 = datasets.make_circles(n_samples=2000, factor=0.5, noise=0.05)
x2, y2 = datasets.make_blobs(n_samples=1000, centers=[[1.2,1.2]], cluster_std=[[.1]])
x = np.concatenate((x1, x2))
plt.scatter(x[:, 0], x[:, 1], marker='o')
plt.show()
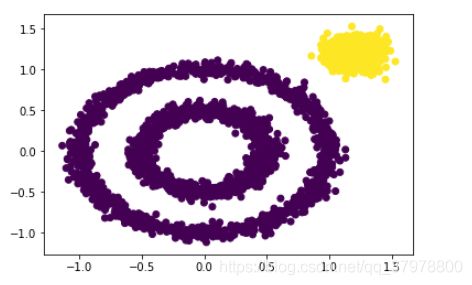
from sklearn.cluster import KMeans
y_pred = KMeans(n_clusters=3).fit_predict(x)
plt.scatter(x[:, 0], x[:, 1], c=y_pred)
plt.show()
from sklearn.cluster import DBSCAN
y_pred = DBSCAN().fit_predict(x)
plt.scatter(x[:, 0], x[:, 1], c=y_pred)
plt.show()
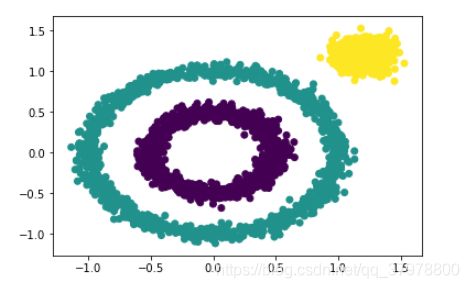
y_pred = DBSCAN(eps = 0.2).fit_predict(x)
plt.scatter(x[:, 0], x[:, 1], c=y_pred)
plt.show()
y_pred = DBSCAN(eps = 0.2, min_samples=50).fit_predict(x)
plt.scatter(x[:, 0], x[:, 1], c=y_pred)
plt.show()