论文地址:https://arxiv.org/abs/1607.00582
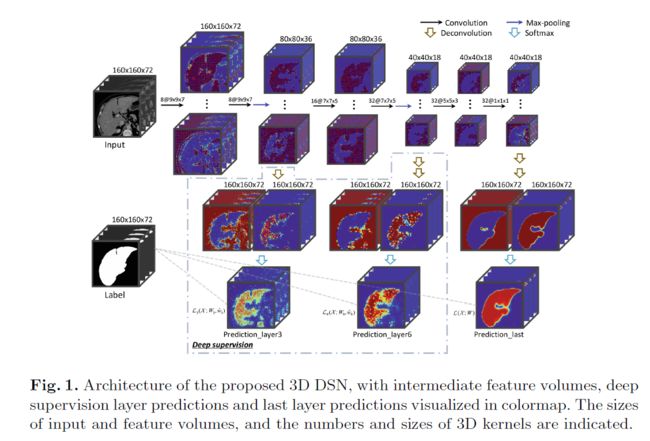
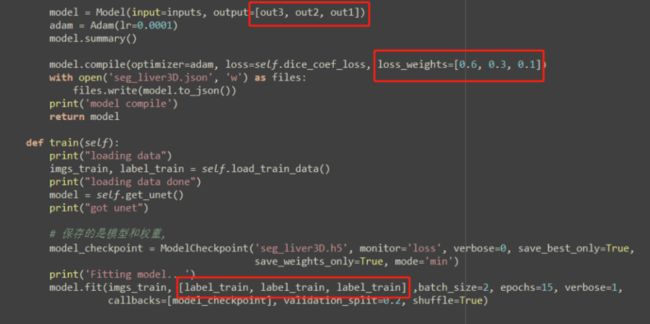
这是一篇MICCAI 2016关于肝脏分割的论文,使用了3D卷积神经网络,难点是虚线里面的部分,如何体现出三个输出的监督作用,最初感觉是在损失函数里面体现出来,尝试把损失函数的一个预测变量改成三个,但运行网络总是报错,后来反复理解这三个输出如何起作用,以及下图中三个输出和label之间的虚线,搜索了与多输出有关的技术博客,最后完全理解了这个网络结构的运行过程,具体到代码的差异性如下红框所示:
模型结构在下采样的过程中有三个输出分支,使用Deconvolution来进行上采样,模型结构比较容易理解,主要是代码实现过程中和平时有一点差异,下图红框是需要注意的地方:
整个网络架构的代码如下:
def DSN(self):
inputs = Input((32,self.img_rows, self.img_cols,1))
conv1 = Conv3D(8, (7, 9, 9), padding='same', activation= 'selu', kernel_initializer = 'he_normal')(inputs)
conv1 = Conv3D(8, (7, 9, 9), padding='same', activation= 'selu',kernel_initializer = 'he_normal')(conv1)
print("conv1 shape:", conv1.shape)
pool1 = MaxPooling3D(pool_size=(2, 2, 2))(conv1)
print("pool1 shape:", pool1.shape)
convT1 = Conv3DTranspose(2, (2, 2, 2), padding='valid', strides = (2, 2, 2), activation= 'selu', kernel_initializer = 'he_normal')(pool1)
print("convT1 shape:", convT1.shape)
out1 = Conv3D(1, 1, activation = 'softmax')(convT1)
conv2 = Conv3D(16, (5, 7, 7), padding='same', activation= 'selu', kernel_initializer = 'he_normal')(pool1)
conv2 = Conv3D(32, (5, 7, 7), padding='same', activation= 'selu',kernel_initializer = 'he_normal')(conv2)
print("conv2 shape:", conv2.shape)
pool2 = MaxPooling3D(pool_size=(2, 2, 2))(conv2)
print("pool2 shape:", pool2.shape)
convT2 = Conv3DTranspose(2, (2, 2, 2), padding='valid', strides = (2, 2, 2), activation= 'selu', kernel_initializer = 'he_normal')(pool2)
convT2 = Conv3DTranspose(2, (2, 2, 2), padding='valid', strides = (2, 2, 2), activation= 'selu', kernel_initializer = 'he_normal')(convT2)
print("convT2 shape:", convT2.shape)
out2 = Conv3D(1, 1, activation = 'softmax')(convT2)
conv3 = Conv3D(32, (3, 5, 5), padding='same', activation= 'selu', kernel_initializer = 'he_normal')(pool2)
conv3 = Conv3D(32, (1, 1, 1), padding='same', activation= 'selu',kernel_initializer = 'he_normal')(conv3)
print("conv3 shape:", conv3.shape)
convT3 = Conv3DTranspose(2, (2, 2, 2), padding='valid', strides = (2, 2, 2), activation= 'selu', kernel_initializer = 'he_normal')(conv3)
convT3 = Conv3DTranspose(2, (2, 2, 2), padding='valid', strides = (2, 2, 2), activation= 'selu', kernel_initializer = 'he_normal')(convT3)
print("convT3 shape:", convT3.shape)
out3 = Conv3D(1, 1, activation = 'sigmoid')(convT3)
model = Model(input=inputs, output=[out3, out2, out1])
adam = Adam(lr=0.0001)
model.summary()
model.compile(optimizer=adam, loss=self.dice_coef_loss, loss_weights=[0.6, 0.3, 0.1])
with open('seg_liver3D.json', 'w') as files:
files.write(model.to_json())
print('model compile')
return model
def train(self):
print("loading data")
imgs_train, label_train = self.load_train_data()
print("loading data done")
model = self.get_unet()
print("got unet")
# 保存的是模型和权重,
model_checkpoint = ModelCheckpoint('seg_liver3D.h5', monitor='loss', verbose=0, save_best_only=True,
save_weights_only=True, mode='min')
print('Fitting model...')
model.fit(imgs_train, [label_train, label_train, label_train] ,batch_size=2, epochs=15, verbose=1,
callbacks=[model_checkpoint], validation_split=0.2, shuffle=True)
网络架构中使用转置卷积来进行上采样过程。