参考文献:https://www.shenxt.info/post/r-circlize-study-2/
1.读取基因表达数据
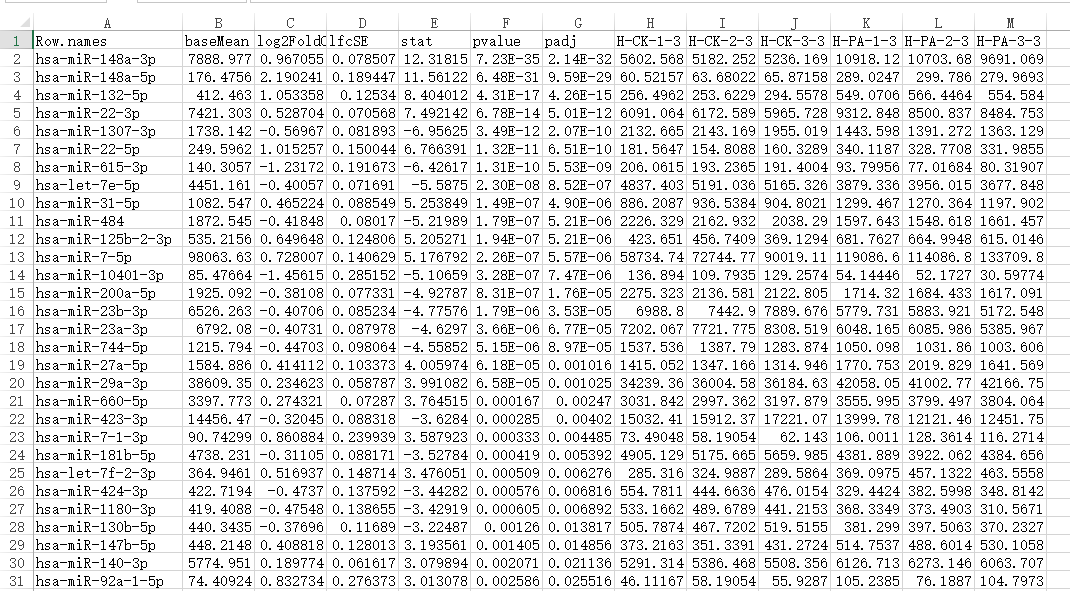
这是我的基因表达量数据
myfiles <- list.files(pattern = "*.csv")
myfiles
matrix<-read.csv(myfiles[1],sep=',',header=T,check.names = FALSE,row.names = 1)
1.1提取部分数据集
matrix<-subset(matrix,padj<0.2)
1.2提取基因表达值所在的列,组成新的矩阵,并将矩阵转置
为什么要转置,参考https://www.jianshu.com/p/115d07af3029
mat =t(matrix[,7:12])
7-12列为每个样本的基因表达量
1.3基因表达归一化
mat=scale(mat, center = TRUE, scale = TRUE)
View(mat)
scale函数可以对矩阵归一化;参考https://www.jianshu.com/p/115d07af3029
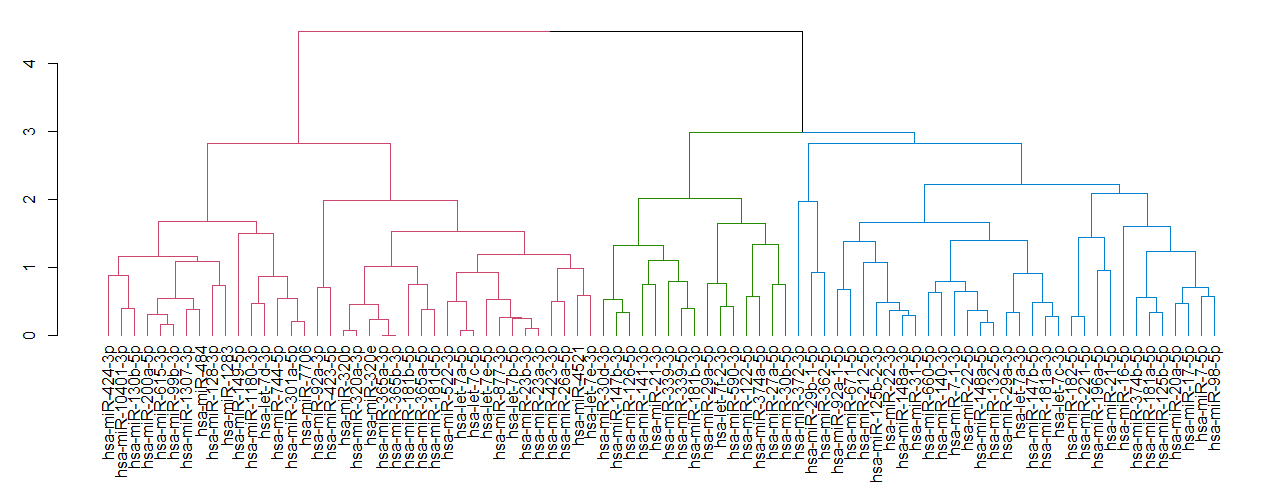
1.4对数据进行聚类,从而得到其dendrogram对象
dend <-as.dendrogram(hclust(dist(t(mat))))
dist函数计算microRNA间的距离,hclust来进行层次聚类
1.5定义进化树颜色
library(dendextend)
n=3
dend <-dend %>% set("branches_k_color", k = n)
n可自定义
1.6可视化
par(mar=c(7.5,3,1,0))
plot(dend)
1.7聚类后的矩阵
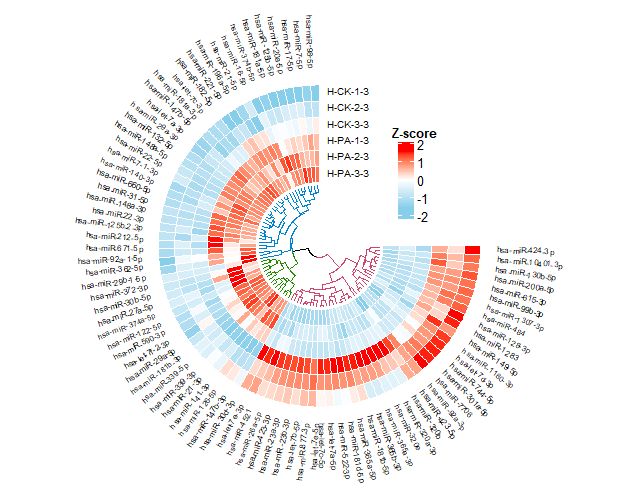
如图Fig 2,聚类后的矩阵的列的顺序会发生变化。按此顺序,重新排列矩阵。
mat2 = mat[, order.dendrogram(dend)]
1.7.1查看矩阵重排后的样本名
lable1=row.names(mat2);lable1
[1] "H-CK-1-3" "H-CK-2-3" "H-CK-3-3" "H-PA-1-3" "H-PA-2-3" "H-PA-3-3"
1.7.2查看矩阵重排后的基因名
lable2=colnames(mat2);lable2
只有基因名顺序,也就是列名顺序发生变化。
nr = nrow(mat2);nr
[1] 6
nc = ncol(mat2);nc
[1] 86
1.8 构建颜色转变函数
require("circlize")
col_fun = colorRamp2(c(-1.5, 0, 1.5), c("skyblue", "white", "red"))
1.9 矩阵中的数值转变为颜色
col_mat = col_fun(mat2)
1.9.1 查看第1列
col_mat[,1]
1.9.1.1 结果
H-CK-1-3 H-CK-2-3 H-CK-3-3 H-PA-1-3 H-PA-2-3 H-PA-3-3
"#FF0000FF" "#FFDED3FF" "#FFAF96FF" "#ABDBF1FF" "#DCF0F9FF" "#BDE3F4FF"
2.1 画板初始化设置
par(mar=c(0,0,0,0))
circos.clear();circos.par(canvas.xlim =c(-1.3,1.3),
canvas.ylim = c(-1.3,1.3),
cell.padding = c(0,0,0,0),
gap.degree =90)
factors = "a"
circos.initialize(factors, xlim = c(0, ncol(mat2)))
xlim和ylim设置1.3,是为了防止添加基因名时溢出
2.2 添加第一个轨道
circos.track(ylim = c(0, nr),bg.border = NA,track.height = 0.1*nr,
panel.fun = function(x, y) {
for(i in 1:nr) {
circos.rect(xleft = 1:nc - 1, ybottom = rep(nr - i, nc),
xright = 1:nc, ytop = rep(nr - i + 1, nc),
border = "white",
col = col_mat[i,])
circos.text(x = nc,
y = 6.4 -i,
labels = lable1[i],
facing = "downward", niceFacing = TRUE,
cex = 0.6,
adj = c(-0.2, 0))
}
})
2.3 添加基因名称
for(i in 1:nc){
circos.text(x = i-0.4,
y = 7,
labels = lable2[i],
facing = "clockwise", niceFacing = TRUE,
cex = 0.4,adj = c(0, 0))
}
2.4 添加进化树
max_height <-max(attr(dend, "height"))
circos.track(ylim = c(0, max_height),bg.border = NA,track.height = 0.3,
panel.fun = function(x, y){
circos.dendrogram(dend = dend,
max_height = max_height)
})
circos.clear()
2.5 添加图例
library(ComplexHeatmap)
lgd <- Legend(at = c(-2,-1, 0, 1, 2), col_fun = col_fun,
title_position = "topcenter",title = "Z-score")
draw(lgd, x = unit(0.7, "npc"), y = unit(0.7, "npc"))